Note
Go to the end to download the full example code
Regression Analysis#
This example uses the diabetes data from sklearn datasets and performs
a regression analysis using a Ridge Regression model. We’ll use the
julearn.PipelineCreator to create a pipeline with two different PCA steps and
reduce the dimensionality of the data, each one computed on a different
subset of features.
# Authors: Georgios Antonopoulos <g.antonopoulos@fz-juelich.de>
# Kaustubh R. Patil <k.patil@fz-juelich.de>
# Shammi More <s.more@fz-juelich.de>
# Federico Raimondo <f.raimondo@fz-juelich.de>
# License: AGPL
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_diabetes
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from julearn import run_cross_validation
from julearn.utils import configure_logging
from julearn.pipeline import PipelineCreator
from julearn.inspect import preprocess
Set the logging level to info to see extra information.
configure_logging(level="INFO")
/home/runner/work/julearn/julearn/julearn/utils/logging.py:66: UserWarning: The '__version__' attribute is deprecated and will be removed in MarkupSafe 3.1. Use feature detection, or `importlib.metadata.version("markupsafe")`, instead.
vstring = str(getattr(module, "__version__", None))
2025-07-07 11:53:18,384 - julearn - INFO - ===== Lib Versions =====
2025-07-07 11:53:18,384 - julearn - INFO - numpy: 1.26.4
2025-07-07 11:53:18,384 - julearn - INFO - scipy: 1.15.3
2025-07-07 11:53:18,384 - julearn - INFO - sklearn: 1.5.2
2025-07-07 11:53:18,384 - julearn - INFO - pandas: 2.2.3
2025-07-07 11:53:18,384 - julearn - INFO - julearn: 0.3.5.dev28
2025-07-07 11:53:18,384 - julearn - INFO - ========================
Load the diabetes data from sklearn as a pandas.DataFrame.
features, target = load_diabetes(return_X_y=True, as_frame=True)
Dataset contains ten variables age, sex, body mass index, average blood pressure, and six blood serum measurements (s1-s6) diabetes patients and a quantitative measure of disease progression one year after baseline which is the target we are interested in predicting.
print("Features: \n", features.head())
print("Target: \n", target.describe())
Features:
age sex bmi ... s4 s5 s6
0 0.038076 0.050680 0.061696 ... -0.002592 0.019907 -0.017646
1 -0.001882 -0.044642 -0.051474 ... -0.039493 -0.068332 -0.092204
2 0.085299 0.050680 0.044451 ... -0.002592 0.002861 -0.025930
3 -0.089063 -0.044642 -0.011595 ... 0.034309 0.022688 -0.009362
4 0.005383 -0.044642 -0.036385 ... -0.002592 -0.031988 -0.046641
[5 rows x 10 columns]
Target:
count 442.000000
mean 152.133484
std 77.093005
min 25.000000
25% 87.000000
50% 140.500000
75% 211.500000
max 346.000000
Name: target, dtype: float64
Let’s combine features and target together in one dataframe and define X and y
Assign types to the features and create feature groups for PCA. We will keep 1 component per PCA group.
X_types = {
"pca1": ["age", "bmi", "bp"],
"pca2": ["s1", "s2", "s3", "s4", "s5", "s6"],
"categorical": ["sex"],
}
Create a pipeline to process the data and the fit a model. We must specify
how each X_type will be used. For example if in the last step we do not
specify apply_to=["continuous", "categorical"], then the pipeline will not
know what to do with the categorical features.
creator = PipelineCreator(problem_type="regression")
creator.add("pca", apply_to="pca1", n_components=1, name="pca_feats1")
creator.add("pca", apply_to="pca2", n_components=1, name="pca_feats2")
creator.add("ridge", apply_to=["continuous", "categorical"])
2025-07-07 11:53:18,399 - julearn - INFO - Adding step pca_feats1 that applies to ColumnTypes<types={'pca1'}; pattern=(?:__:type:__pca1)>
2025-07-07 11:53:18,399 - julearn - INFO - Setting hyperparameter n_components = 1
2025-07-07 11:53:18,399 - julearn - INFO - Step added
2025-07-07 11:53:18,399 - julearn - INFO - Adding step pca_feats2 that applies to ColumnTypes<types={'pca2'}; pattern=(?:__:type:__pca2)>
2025-07-07 11:53:18,399 - julearn - INFO - Setting hyperparameter n_components = 1
2025-07-07 11:53:18,399 - julearn - INFO - Step added
2025-07-07 11:53:18,400 - julearn - INFO - Adding step ridge that applies to ColumnTypes<types={'continuous', 'categorical'}; pattern=(?:__:type:__continuous|__:type:__categorical)>
2025-07-07 11:53:18,400 - julearn - INFO - Step added
<julearn.pipeline.pipeline_creator.PipelineCreator object at 0x7f86f5f1a980>
Split the dataset into train and test.
train_diabetes, test_diabetes = train_test_split(data_diabetes, test_size=0.3)
Train a ridge regression model on train dataset and use mean absolute error for scoring.
2025-07-07 11:53:18,401 - julearn - INFO - ==== Input Data ====
2025-07-07 11:53:18,401 - julearn - INFO - Using dataframe as input
2025-07-07 11:53:18,401 - julearn - INFO - Features: ['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
2025-07-07 11:53:18,401 - julearn - INFO - Target: target
2025-07-07 11:53:18,401 - julearn - INFO - Expanded features: ['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
2025-07-07 11:53:18,401 - julearn - INFO - X_types:{'pca1': ['age', 'bmi', 'bp'], 'pca2': ['s1', 's2', 's3', 's4', 's5', 's6'], 'categorical': ['sex']}
2025-07-07 11:53:18,402 - julearn - INFO - ====================
2025-07-07 11:53:18,402 - julearn - INFO -
2025-07-07 11:53:18,403 - julearn - INFO - = Model Parameters =
2025-07-07 11:53:18,403 - julearn - INFO - ====================
2025-07-07 11:53:18,403 - julearn - INFO -
2025-07-07 11:53:18,403 - julearn - INFO - = Data Information =
2025-07-07 11:53:18,403 - julearn - INFO - Problem type: regression
2025-07-07 11:53:18,403 - julearn - INFO - Number of samples: 309
2025-07-07 11:53:18,403 - julearn - INFO - Number of features: 10
2025-07-07 11:53:18,403 - julearn - INFO - ====================
2025-07-07 11:53:18,403 - julearn - INFO -
2025-07-07 11:53:18,404 - julearn - INFO - Target type: float64
2025-07-07 11:53:18,404 - julearn - INFO - Using outer CV scheme KFold(n_splits=5, random_state=None, shuffle=False) (incl. final model)
The scores dataframe has all the values for each CV split.
print(scores.head())
fit_time score_time ... fold cv_mdsum
0 0.013479 0.006198 ... 0 b10eef89b4192178d482d7a1587a248a
1 0.013615 0.006193 ... 1 b10eef89b4192178d482d7a1587a248a
2 0.013362 0.006165 ... 2 b10eef89b4192178d482d7a1587a248a
3 0.013562 0.006153 ... 3 b10eef89b4192178d482d7a1587a248a
4 0.013388 0.006193 ... 4 b10eef89b4192178d482d7a1587a248a
[5 rows x 8 columns]
Mean value of mean absolute error across CV.
print(scores["test_score"].mean())
0.3107976743678922
Let’s see how the data looks like after preprocessing. We will process the data until the first PCA step. We should get the first PCA component for [“age”, “bmi”, “bp”] and leave other features untouched.
data_processed1 = preprocess(model, X, data=train_diabetes, until="pca_feats1")
print("Data after preprocessing until PCA step 1")
data_processed1.head()
Data after preprocessing until PCA step 1
We will process the data until the second PCA step. We should now also get one PCA component for [“s1”, “s2”, “s3”, “s4”, “s5”, “s6”].
data_processed2 = preprocess(model, X, data=train_diabetes, until="pca_feats2")
print("Data after preprocessing until PCA step 2")
data_processed2.head()
Data after preprocessing until PCA step 2
Now we can get the MAE fold and repetition:
df_mae = scores.set_index(["repeat", "fold"])["test_score"].unstack() * -1
df_mae.index.name = "Repeats"
df_mae.columns.name = "K-fold splits"
print(df_mae)
K-fold splits 0 1 2 3 4
Repeats
0 -0.341472 -0.348168 -0.269257 -0.286067 -0.309025
Plot heatmap of mean absolute error (MAE) over all repeats and CV splits.
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.heatmap(df_mae, cmap="YlGnBu")
plt.title("Cross-validation MAE")
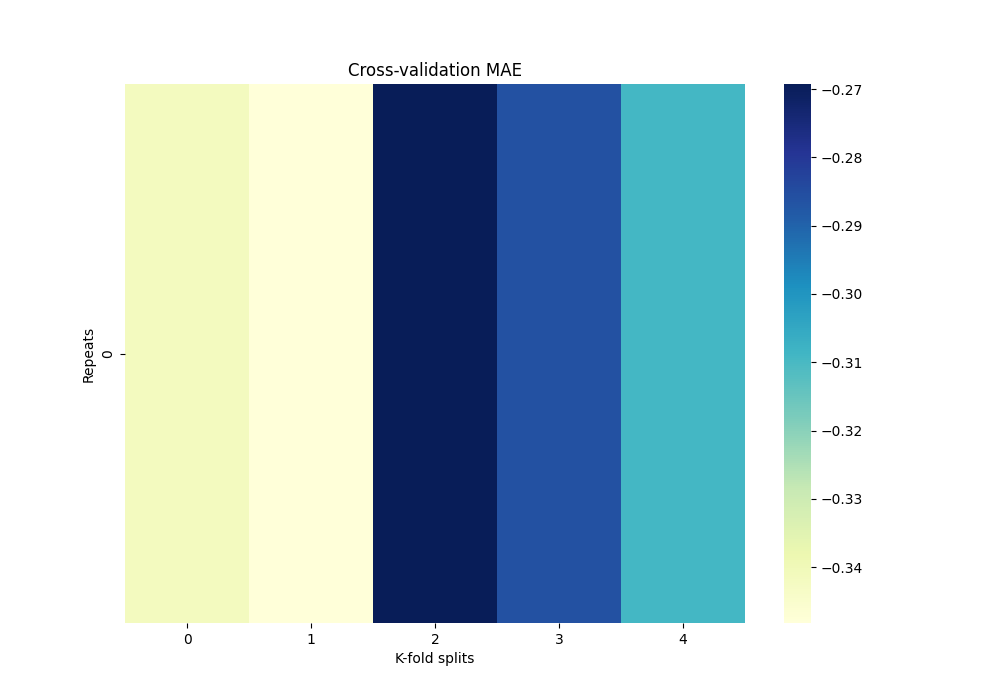
Text(0.5, 1.0, 'Cross-validation MAE')
Use the final model to make predictions on test data and plot scatterplot of true values vs predicted values.
y_true = test_diabetes[y]
y_pred = model.predict(test_diabetes[X])
mae = format(mean_absolute_error(y_true, y_pred), ".2f")
corr = format(np.corrcoef(y_pred, y_true)[1, 0], ".2f")
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.set_style("darkgrid")
plt.scatter(y_true, y_pred)
plt.plot(y_true, y_true)
xmin, xmax = ax.get_xlim()
ymin, ymax = ax.get_ylim()
text = "MAE: " + str(mae) + " CORR: " + str(corr)
ax.set(xlabel="True values", ylabel="Predicted values")
plt.title("Actual vs Predicted")
plt.text(
xmax - 0.01 * xmax,
ymax - 0.01 * ymax,
text,
verticalalignment="top",
horizontalalignment="right",
fontsize=12,
)
plt.axis("scaled")
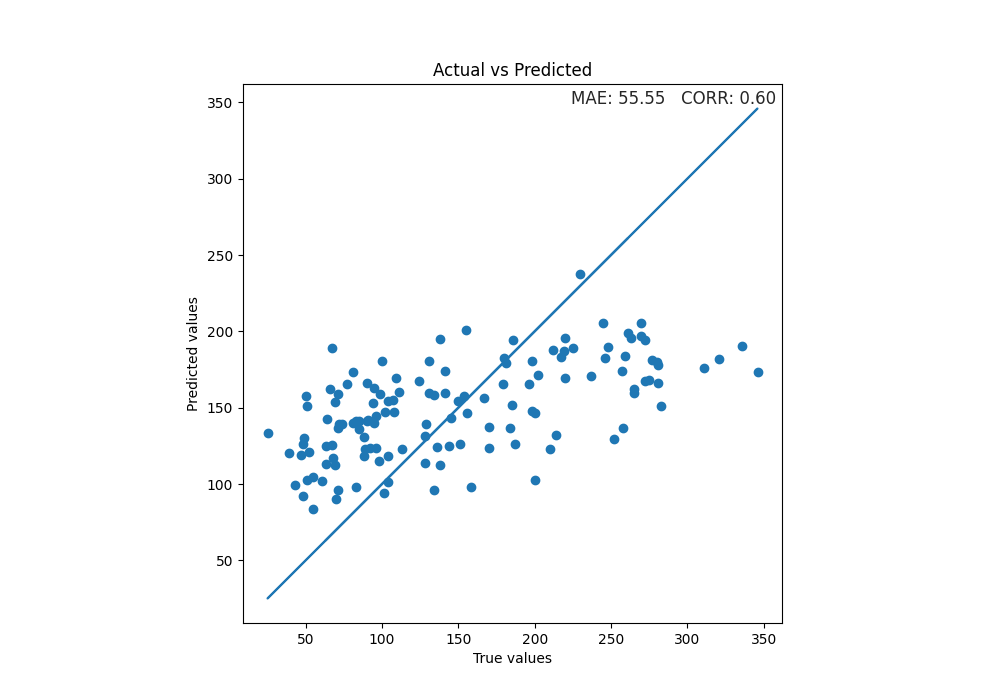
(8.95, 362.05, 8.95, 362.05)
Total running time of the script: (0 minutes 0.372 seconds)