Note
Go to the end to download the full example code
Inspecting Random Forest models#
This example uses the ‘iris’ dataset, performs simple binary classification using a Random Forest classifier and analyse the model.
# Authors: Federico Raimondo <f.raimondo@fz-juelich.de>
#
# License: AGPL
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from seaborn import load_dataset
from julearn import run_cross_validation
from julearn.utils import configure_logging
Set the logging level to info to see extra information
configure_logging(level="INFO")
2023-07-19 12:41:53,802 - julearn - INFO - ===== Lib Versions =====
2023-07-19 12:41:53,802 - julearn - INFO - numpy: 1.25.1
2023-07-19 12:41:53,802 - julearn - INFO - scipy: 1.11.1
2023-07-19 12:41:53,802 - julearn - INFO - sklearn: 1.3.0
2023-07-19 12:41:53,802 - julearn - INFO - pandas: 2.0.3
2023-07-19 12:41:53,802 - julearn - INFO - julearn: 0.3.1.dev1
2023-07-19 12:41:53,802 - julearn - INFO - ========================
Random Forest variable importance#
df_iris = load_dataset("iris")
The dataset has three kind of species. We will keep two to perform a binary classification.
We will use a Random Forest classifier. By setting
return_estimator=’final’, the run_cross_validation() function
returns the estimator fitted with all the data.
2023-07-19 12:41:53,806 - julearn - INFO - ==== Input Data ====
2023-07-19 12:41:53,806 - julearn - INFO - Using dataframe as input
2023-07-19 12:41:53,806 - julearn - INFO - Features: ['sepal_length', 'sepal_width', 'petal_length']
2023-07-19 12:41:53,806 - julearn - INFO - Target: species
2023-07-19 12:41:53,806 - julearn - INFO - Expanded features: ['sepal_length', 'sepal_width', 'petal_length']
2023-07-19 12:41:53,806 - julearn - INFO - X_types:{}
2023-07-19 12:41:53,806 - julearn - WARNING - The following columns are not defined in X_types: ['sepal_length', 'sepal_width', 'petal_length']. They will be treated as continuous.
/home/runner/work/julearn/julearn/julearn/utils/logging.py:238: RuntimeWarning: The following columns are not defined in X_types: ['sepal_length', 'sepal_width', 'petal_length']. They will be treated as continuous.
warn(msg, category=category)
2023-07-19 12:41:53,807 - julearn - INFO - ====================
2023-07-19 12:41:53,807 - julearn - INFO -
2023-07-19 12:41:53,807 - julearn - INFO - Adding step zscore that applies to ColumnTypes<types={'continuous'}; pattern=(?:__:type:__continuous)>
2023-07-19 12:41:53,807 - julearn - INFO - Step added
2023-07-19 12:41:53,807 - julearn - INFO - Adding step rf that applies to ColumnTypes<types={'continuous'}; pattern=(?:__:type:__continuous)>
2023-07-19 12:41:53,807 - julearn - INFO - Step added
2023-07-19 12:41:53,808 - julearn - INFO - = Model Parameters =
2023-07-19 12:41:53,808 - julearn - INFO - ====================
2023-07-19 12:41:53,808 - julearn - INFO -
2023-07-19 12:41:53,808 - julearn - INFO - = Data Information =
2023-07-19 12:41:53,808 - julearn - INFO - Problem type: classification
2023-07-19 12:41:53,808 - julearn - INFO - Number of samples: 100
2023-07-19 12:41:53,808 - julearn - INFO - Number of features: 3
2023-07-19 12:41:53,808 - julearn - INFO - ====================
2023-07-19 12:41:53,808 - julearn - INFO -
2023-07-19 12:41:53,809 - julearn - INFO - Number of classes: 2
2023-07-19 12:41:53,809 - julearn - INFO - Target type: object
2023-07-19 12:41:53,809 - julearn - INFO - Class distributions: species
versicolor 50
virginica 50
Name: count, dtype: int64
2023-07-19 12:41:53,810 - julearn - INFO - Using outer CV scheme KFold(n_splits=5, random_state=None, shuffle=False)
2023-07-19 12:41:53,810 - julearn - INFO - Binary classification problem detected.
This type of classifier has an internal variable that can inform us on how
_important_ is each of the features. Caution: read the proper scikit-learn
documentation RandomForestClassifier to understandhow this learning algorithm works.
rf = model_iris["rf"]
to_plot = pd.DataFrame(
{
"variable": [x.replace("_", " ") for x in X],
"importance": rf.feature_importances_,
}
)
fig, ax = plt.subplots(1, 1, figsize=(6, 4))
sns.barplot(x="importance", y="variable", data=to_plot, ax=ax)
ax.set_title("Variable Importances for Random Forest Classifier")
fig.tight_layout()
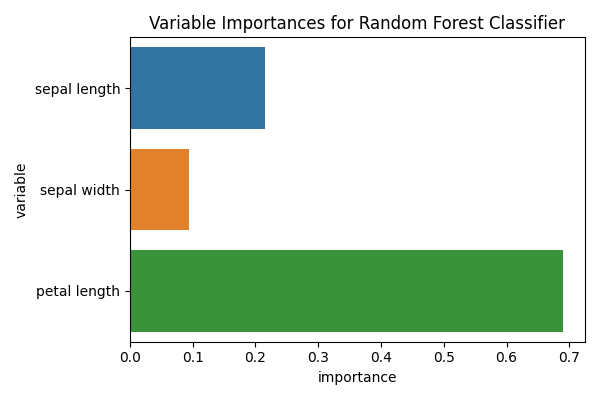
However, some reviewers (including myself), might wander about the variability of the importance of these features. In the previous example all the feature importances were obtained by fitting on the entire dataset, while the performance was estimated using cross validation.
By specifying return_estimator=’cv’, we can get, for each fold, the fitted estimator.
2023-07-19 12:41:54,597 - julearn - INFO - ==== Input Data ====
2023-07-19 12:41:54,598 - julearn - INFO - Using dataframe as input
2023-07-19 12:41:54,598 - julearn - INFO - Features: ['sepal_length', 'sepal_width', 'petal_length']
2023-07-19 12:41:54,598 - julearn - INFO - Target: species
2023-07-19 12:41:54,598 - julearn - INFO - Expanded features: ['sepal_length', 'sepal_width', 'petal_length']
2023-07-19 12:41:54,598 - julearn - INFO - X_types:{}
2023-07-19 12:41:54,598 - julearn - WARNING - The following columns are not defined in X_types: ['sepal_length', 'sepal_width', 'petal_length']. They will be treated as continuous.
/home/runner/work/julearn/julearn/julearn/utils/logging.py:238: RuntimeWarning: The following columns are not defined in X_types: ['sepal_length', 'sepal_width', 'petal_length']. They will be treated as continuous.
warn(msg, category=category)
2023-07-19 12:41:54,599 - julearn - INFO - ====================
2023-07-19 12:41:54,599 - julearn - INFO -
2023-07-19 12:41:54,599 - julearn - INFO - Adding step zscore that applies to ColumnTypes<types={'continuous'}; pattern=(?:__:type:__continuous)>
2023-07-19 12:41:54,599 - julearn - INFO - Step added
2023-07-19 12:41:54,599 - julearn - INFO - Adding step rf that applies to ColumnTypes<types={'continuous'}; pattern=(?:__:type:__continuous)>
2023-07-19 12:41:54,599 - julearn - INFO - Step added
2023-07-19 12:41:54,600 - julearn - INFO - = Model Parameters =
2023-07-19 12:41:54,600 - julearn - INFO - ====================
2023-07-19 12:41:54,600 - julearn - INFO -
2023-07-19 12:41:54,600 - julearn - INFO - = Data Information =
2023-07-19 12:41:54,600 - julearn - INFO - Problem type: classification
2023-07-19 12:41:54,600 - julearn - INFO - Number of samples: 100
2023-07-19 12:41:54,600 - julearn - INFO - Number of features: 3
2023-07-19 12:41:54,600 - julearn - INFO - ====================
2023-07-19 12:41:54,601 - julearn - INFO -
2023-07-19 12:41:54,601 - julearn - INFO - Number of classes: 2
2023-07-19 12:41:54,601 - julearn - INFO - Target type: object
2023-07-19 12:41:54,601 - julearn - INFO - Class distributions: species
versicolor 50
virginica 50
Name: count, dtype: int64
2023-07-19 12:41:54,602 - julearn - INFO - Using outer CV scheme KFold(n_splits=5, random_state=None, shuffle=False)
2023-07-19 12:41:54,602 - julearn - INFO - Binary classification problem detected.
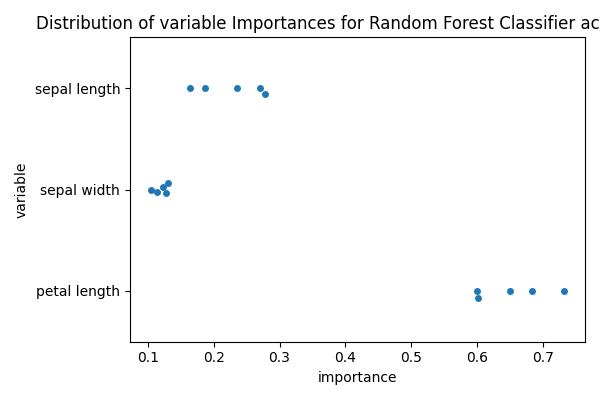
Now we can obtain the feature importance for each estimator (CV fold)
to_plot = []
for i_fold, estimator in enumerate(scores["estimator"]):
this_importances = pd.DataFrame(
{
"variable": [x.replace("_", " ") for x in X],
"importance": estimator["rf"].feature_importances_,
"fold": i_fold,
}
)
to_plot.append(this_importances)
to_plot = pd.concat(to_plot)
Finally, we can plot the variable importances for each fold
Total running time of the script: ( 0 minutes 1.562 seconds)