Note
Go to the end to download the full example code
Preprocessing with variance threshold, zscore and PCA
This example uses the ‘iris’ dataset, performs simple binary classification after the pre-processing the features including removal of low variance features, feature normalization using zscore and feature reduction using PCA. We will check the features after each preprocessing step.
# Authors: Shammi More <s.more@fz-juelich.de>
#
# License: AGPL
import matplotlib.pyplot as plt
import seaborn as sns
from seaborn import load_dataset
from julearn import run_cross_validation
from julearn.utils import configure_logging
Set the logging level to info to see extra information
configure_logging(level='INFO')
2023-04-06 09:51:01,275 - julearn - INFO - ===== Lib Versions =====
2023-04-06 09:51:01,275 - julearn - INFO - numpy: 1.23.5
2023-04-06 09:51:01,275 - julearn - INFO - scipy: 1.10.1
2023-04-06 09:51:01,275 - julearn - INFO - sklearn: 1.0.2
2023-04-06 09:51:01,275 - julearn - INFO - pandas: 1.4.4
2023-04-06 09:51:01,276 - julearn - INFO - julearn: 0.3.1.dev2
2023-04-06 09:51:01,276 - julearn - INFO - ========================
Load the iris data from seaborn
df_iris = load_dataset('iris')
The dataset has three kind of species. We will keep two to perform a binary classification.
df_iris = df_iris[df_iris['species'].isin(['versicolor', 'virginica'])]
We will use the sepal length, width and petal length and petal width as features and predict the species
Let’s look at the summary statistics of the raw features
print('Summary Statistics of the raw features : \n', df_iris.describe())
Summary Statistics of the raw features :
sepal_length sepal_width petal_length petal_width
count 100.000000 100.000000 100.000000 100.000000
mean 6.262000 2.872000 4.906000 1.676000
std 0.662834 0.332751 0.825578 0.424769
min 4.900000 2.000000 3.000000 1.000000
25% 5.800000 2.700000 4.375000 1.300000
50% 6.300000 2.900000 4.900000 1.600000
75% 6.700000 3.025000 5.525000 2.000000
max 7.900000 3.800000 6.900000 2.500000
We will preprocess the features using variance thresholding, zscore and PCA and then train a random forest model
# Define the model parameters and preprocessing steps first
# Setting the threshold for variance to 0.15, number of PCA components to 2
# and number of trees for random forest to 200
model_params = {'select_variance__threshold': 0.15,
'pca__n_components': 2,
'rf__n_estimators': 200}
preprocess_X = ['select_variance', 'zscore', 'pca']
scores, model = run_cross_validation(
X=X, y=y, data=df_iris, model='rf', preprocess_X=preprocess_X,
scoring=['accuracy', 'roc_auc'], model_params=model_params,
return_estimator='final', seed=200)
2023-04-06 09:51:01,299 - julearn - INFO - Setting random seed to 200
2023-04-06 09:51:01,299 - julearn - INFO - Using default CV
2023-04-06 09:51:01,299 - julearn - INFO - ==== Input Data ====
2023-04-06 09:51:01,299 - julearn - INFO - Using dataframe as input
2023-04-06 09:51:01,300 - julearn - INFO - Features: ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
2023-04-06 09:51:01,300 - julearn - INFO - Target: species
2023-04-06 09:51:01,300 - julearn - INFO - Expanded X: ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
2023-04-06 09:51:01,300 - julearn - INFO - Expanded Confounds: []
2023-04-06 09:51:01,301 - julearn - INFO - ====================
2023-04-06 09:51:01,301 - julearn - INFO -
2023-04-06 09:51:01,301 - julearn - INFO - ====== Model ======
2023-04-06 09:51:01,301 - julearn - INFO - Obtaining model by name: rf
2023-04-06 09:51:01,302 - julearn - INFO - ===================
2023-04-06 09:51:01,302 - julearn - INFO -
2023-04-06 09:51:01,302 - julearn - INFO - = Model Parameters =
2023-04-06 09:51:01,302 - julearn - INFO - Setting hyperparameter select_variance__threshold = 0.15
2023-04-06 09:51:01,303 - julearn - INFO - Setting hyperparameter pca__n_components = 2
2023-04-06 09:51:01,304 - julearn - INFO - Setting hyperparameter rf__n_estimators = 200
2023-04-06 09:51:01,306 - julearn - INFO - ====================
2023-04-06 09:51:01,306 - julearn - INFO -
2023-04-06 09:51:01,306 - julearn - INFO - CV interpreted as RepeatedKFold with 5 repetitions of 5 folds
Now let’s look at the data after pre-processing. It can be done using ‘preprocess’ method. By default it will apply all the pre-processing steps (‘select_variance’, ‘zscore’, ‘pca’ in this case) and return pre-processed data. Note that here we are applying pre-processing only on X. Notice that the column names have changed in this new dataframe.
pre_X, pre_y = model.preprocess(df_iris[X], df_iris[y])
print('Features after PCA : \n', pre_X)
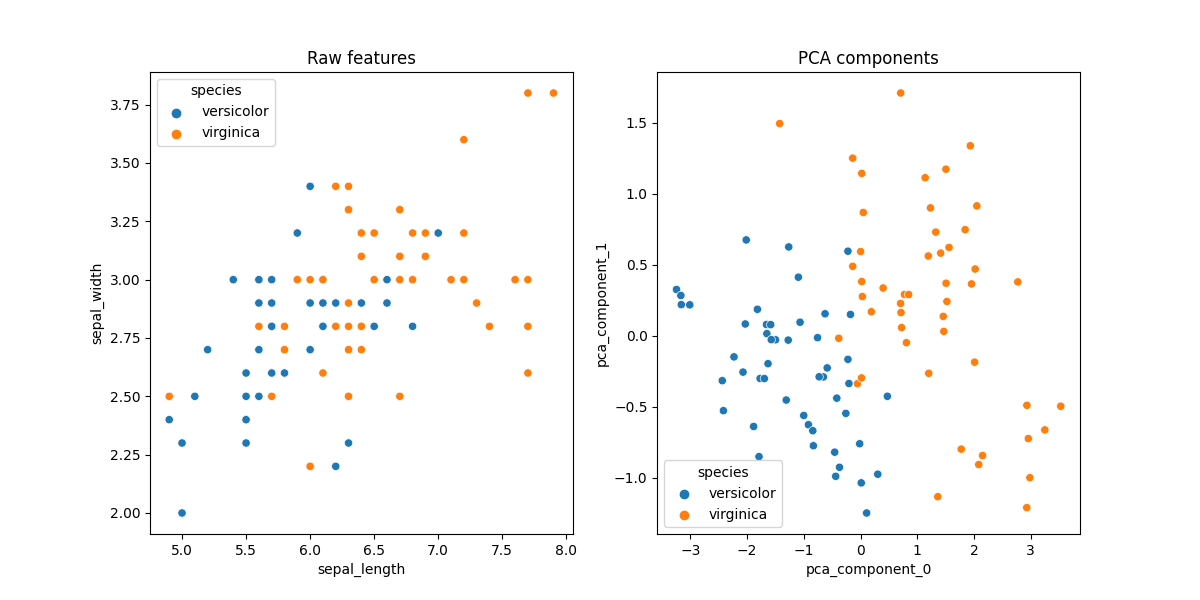
# Let's plot scatter plots for raw features and the PCA components
pre_df = pre_X.join(pre_y)
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
sns.scatterplot(x='sepal_length', y='sepal_width', data=df_iris, hue='species',
ax=axes[0])
axes[0].set_title('Raw features')
sns.scatterplot(x='pca_component_0', y='pca_component_1', data=pre_df,
hue='species', ax=axes[1])
axes[1].set_title('PCA components')
Features after PCA :
pca_component_0 pca_component_1
50 0.107487 -1.249219
51 -0.418588 -0.440114
52 0.304093 -0.975911
53 -1.818796 0.185820
54 -0.259106 -0.547432
.. ... ...
145 1.413947 0.581755
146 0.397898 0.335935
147 0.848759 0.289674
148 1.139654 1.112779
149 0.001729 0.592856
[100 rows x 2 columns]
Text(0.5, 1.0, 'PCA components')
But let’s say we want to look at features after applying only one or more preprocessing steps eg: only variance thresholding or till zscore. To do so we can set the argument until to the desired preprocessing step. Note that the name of the preprocessing step is the same as used in the run_cross_validation function in preprocess_X.
# Let's look at features after variance thresholding. We see that now we have
# one feature less as the variance for this feature ('sepal_width') was below
# the set threshold.
var_th_X, var_th_y = model.preprocess(df_iris[X], df_iris[y],
until='select_variance')
print('Features after variance thresholding: \n', var_th_X)
Features after variance thresholding:
sepal_length petal_length petal_width
50 7.0 4.7 1.4
51 6.4 4.5 1.5
52 6.9 4.9 1.5
53 5.5 4.0 1.3
54 6.5 4.6 1.5
.. ... ... ...
145 6.7 5.2 2.3
146 6.3 5.0 1.9
147 6.5 5.2 2.0
148 6.2 5.4 2.3
149 5.9 5.1 1.8
[100 rows x 3 columns]
Now let’s see features after variance thresholding and zscoring. We can now set the until argument to ‘zscore’
Let’s look at the summary statistics of the zscored features. We see here that the mean of all the features is zero and standard deviation is one.
print('Summary Statistics of the zscored features : \n', zscored_df.describe())
Summary Statistics of the zscored features :
sepal_length petal_length petal_width
count 1.000000e+02 1.000000e+02 1.000000e+02
mean -6.683543e-16 3.269607e-16 -1.443845e-15
std 1.005038e+00 1.005038e+00 1.005038e+00
min -2.065164e+00 -2.320315e+00 -1.599473e+00
25% -7.005180e-01 -6.464256e-01 -8.896475e-01
50% 5.761837e-02 -7.304244e-03 -1.798224e-01
75% 6.641275e-01 7.535546e-01 7.666111e-01
max 2.483655e+00 2.427444e+00 1.949653e+00
We can also look at the features pre-processed till PCA. Since ‘pca’ is the last preprocessing step we don’t really need the until argument (as shown above).
Features after PCA :
pca_component_0 pca_component_1
50 0.107487 -1.249219
51 -0.418588 -0.440114
52 0.304093 -0.975911
53 -1.818796 0.185820
54 -0.259106 -0.547432
.. ... ...
145 1.413947 0.581755
146 0.397898 0.335935
147 0.848759 0.289674
148 1.139654 1.112779
149 0.001729 0.592856
[100 rows x 2 columns]
Total running time of the script: ( 0 minutes 11.272 seconds)