Note
Go to the end to download the full example code
Multiclass Classification
This example uses the ‘iris’ dataset and performs multiclass classification using a Support Vector Machine classifier and plots heatmaps for cross-validation accuracies and plots confusion matrix for the test data.
# Authors: Shammi More <s.more@fz-juelich.de>
# Federico Raimondo <f.raimondo@fz-juelich.de>
#
# License: AGPL
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from seaborn import load_dataset
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from julearn import run_cross_validation
from julearn.utils import configure_logging
Set the logging level to info to see extra information
2023-04-06 09:50:57,312 - julearn - INFO - ===== Lib Versions =====
2023-04-06 09:50:57,312 - julearn - INFO - numpy: 1.23.5
2023-04-06 09:50:57,312 - julearn - INFO - scipy: 1.10.1
2023-04-06 09:50:57,312 - julearn - INFO - sklearn: 1.0.2
2023-04-06 09:50:57,312 - julearn - INFO - pandas: 1.4.4
2023-04-06 09:50:57,312 - julearn - INFO - julearn: 0.3.1.dev2
2023-04-06 09:50:57,313 - julearn - INFO - ========================
Split the dataset into train and test
train_iris, test_iris = train_test_split(df_iris, test_size=0.2,
stratify=df_iris[y])
Perform multiclass classification as iris dataset contains 3 kinds of species
2023-04-06 09:50:57,317 - julearn - INFO - Using default CV
2023-04-06 09:50:57,317 - julearn - INFO - ==== Input Data ====
2023-04-06 09:50:57,317 - julearn - INFO - Using dataframe as input
2023-04-06 09:50:57,318 - julearn - INFO - Features: ['sepal_length', 'sepal_width', 'petal_length']
2023-04-06 09:50:57,318 - julearn - INFO - Target: species
2023-04-06 09:50:57,318 - julearn - INFO - Expanded X: ['sepal_length', 'sepal_width', 'petal_length']
2023-04-06 09:50:57,318 - julearn - INFO - Expanded Confounds: []
2023-04-06 09:50:57,319 - julearn - INFO - ====================
2023-04-06 09:50:57,319 - julearn - INFO -
2023-04-06 09:50:57,319 - julearn - INFO - ====== Model ======
2023-04-06 09:50:57,319 - julearn - INFO - Obtaining model by name: svm
2023-04-06 09:50:57,319 - julearn - INFO - ===================
2023-04-06 09:50:57,319 - julearn - INFO -
2023-04-06 09:50:57,319 - julearn - INFO - CV interpreted as RepeatedKFold with 5 repetitions of 5 folds
The scores dataframe has all the values for each CV split.
print(scores.head())
fit_time score_time test_accuracy repeat fold
0 0.012913 0.008849 0.875000 0 0
1 0.012830 0.008488 0.916667 0 1
2 0.012852 0.008507 0.875000 0 2
3 0.012333 0.008412 0.875000 0 3
4 0.012681 0.008568 1.000000 0 4
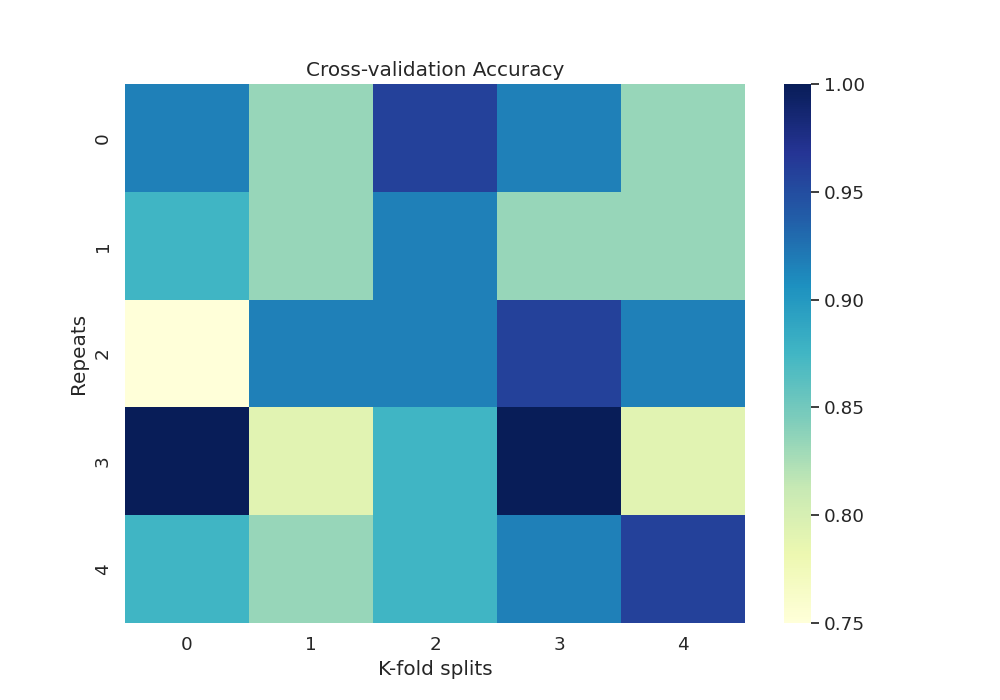
Now we can get the accuracy per fold and repetition:
df_accuracy = scores.set_index(
['repeat', 'fold'])['test_accuracy'].unstack()
df_accuracy.index.name = 'Repeats'
df_accuracy.columns.name = 'K-fold splits'
print(df_accuracy)
K-fold splits 0 1 2 3 4
Repeats
0 0.875000 0.916667 0.875000 0.875000 1.000000
1 0.958333 0.916667 0.833333 0.875000 0.958333
2 0.958333 0.958333 0.916667 0.916667 0.916667
3 0.833333 0.916667 0.875000 0.958333 0.958333
4 1.000000 0.875000 0.833333 0.916667 0.875000
Plot heatmap of accuracy over all repeats and CV splits
sns.set(font_scale=1.2)
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.heatmap(df_accuracy, cmap="YlGnBu")
plt.title('Cross-validation Accuracy')
Text(0.5, 1.0, 'Cross-validation Accuracy')
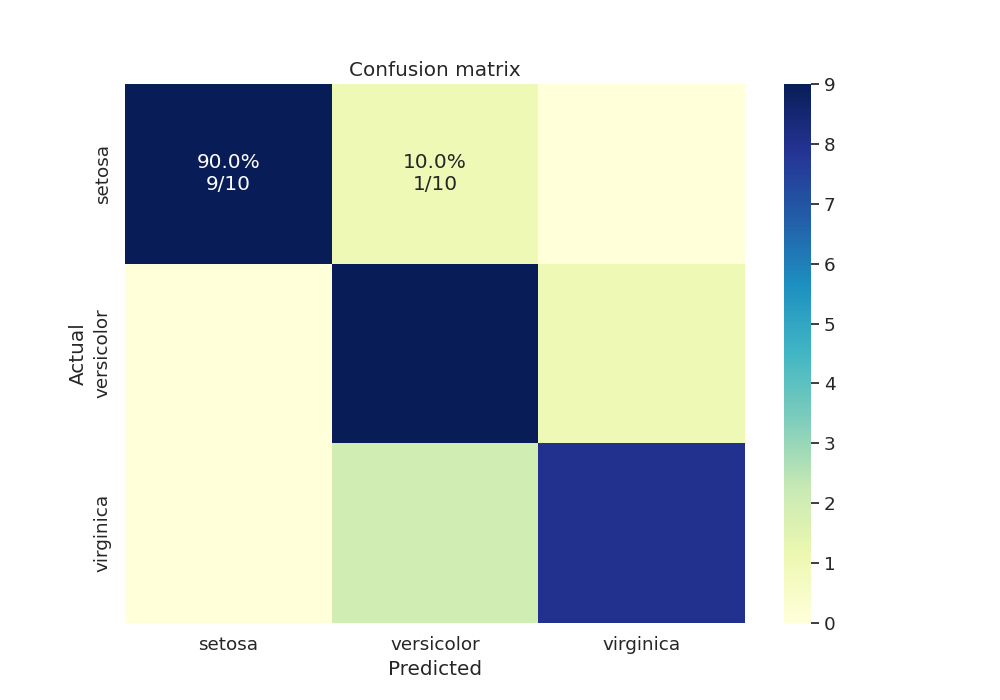
We can also test our final model’s accuracy and plot the confusion matrix for the test data as an annotated heatmap
y_true = test_iris[y]
y_pred = model_iris.predict(test_iris[X])
cm = confusion_matrix(y_true, y_pred, labels=np.unique(y_true))
print(cm)
[[10 0 0]
[ 0 10 0]
[ 0 1 9]]
Now that we have our confusion matrix, let’s build another matrix with annotations.
cm_sum = np.sum(cm, axis=1, keepdims=True)
cm_perc = cm / cm_sum.astype(float) * 100
annot = np.empty_like(cm).astype(str)
nrows, ncols = cm.shape
for i in range(nrows):
for j in range(ncols):
c = cm[i, j]
p = cm_perc[i, j]
if c == 0:
annot[i, j] = ''
else:
s = cm_sum[i]
annot[i, j] = '%.1f%%\n%d/%d' % (p, c, s)
Finally we create another dataframe with the confusion matrix and plot the heatmap with annotations.
cm = pd.DataFrame(cm, index=np.unique(y_true), columns=np.unique(y_true))
cm.index.name = 'Actual'
cm.columns.name = 'Predicted'
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.heatmap(cm, cmap="YlGnBu", annot=annot, fmt='', ax=ax)
plt.title('Confusion matrix')
Text(0.5, 1.0, 'Confusion matrix')
Total running time of the script: ( 0 minutes 1.099 seconds)