Analysis 3c: Introspect: Real World Datasets Audio Data Features Importance
Analysis 3c: Introspect: Real World Datasets Audio Data Features Importance¶
Imports, Functs and Paths
from sklearn.inspection import DecisionBoundaryDisplay
from sklearn.tree import DecisionTreeClassifier
from myst_nb import glue
from sklearn.preprocessing import MinMaxScaler
from julearn import run_cross_validation
import joblib
import os
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# plot styles
from sciplotlib import style
import matplotlib as mpl
from matplotlib.colors import LinearSegmentedColormap
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
base_save_paper = "./paper_val/"
results_base = "../../results/permutations"
base_dir = "../../"
colors = [
"#E64B35",
"#4DBBD5",
"#00A087",
"#3C5488",
"#F39B7F",
"#8491B4",
"#91D1C2FF",
"#DC0000",
"#7E6148",
"#B09C85",
]
red = colors[0]
blue = colors[1]
green = colors[2]
purple = colors[5]
def mm_to_inch(val_in_inch):
mm = 0.1 / 2.54
return val_in_inch * mm
base_save_paper = "./paper_val/"
results_base = "../../results/"
base_dir = "../../"
audio_data_non_TaCo_folder = f"{results_base}basic_non_TaCo/realistic_non_TaCo/models/"
audio_data_shuffled_non_TaCo_folder = (
f"{results_base}" "shuffled_features_non_TaCo/realistic_non_TaCo/models/"
)
mpl.style.use(style.get_style("nature-reviews"))
mpl.rc("xtick", labelsize=11)
mpl.rc("ytick", labelsize=11)
mpl.rc("axes", labelsize=12, titlesize=12)
mpl.rc("figure", dpi=300)
mpl.rc("figure.subplot", wspace=mm_to_inch(8), hspace=0.7)
mpl.rc("lines", linewidth=1, markersize=2)
fig = plt.figure(
figsize=[mm_to_inch(183), mm_to_inch(140)],
)
<Figure size 2161.42x1653.54 with 0 Axes>
Some info on the data:
os.listdir(audio_data_non_TaCo_folder)
df_original = pd.read_csv("../../data/realistic_non_TaCo/audio_data/audio_data_BDI.csv")
# Get the models ready and import data:
X = df_original.filter(regex=".*__continuous$").columns.tolist()
y = "ATT_Task__binary_target"
confounds = ["BDI__continuous_confound"]
model_raw = joblib.load(
audio_data_non_TaCo_folder + "audio_data_BDI___rf___False___716845___BDI.joblib"
)
model_rem = joblib.load(
audio_data_non_TaCo_folder + "audio_data_BDI___rf___True___716845___BDI.joblib"
)
model_raw_shuffled = joblib.load(
audio_data_shuffled_non_TaCo_folder
+ "audio_data_BDI___rf___False___716845___BDI.joblib"
)
model_rem_shuffled = joblib.load(
audio_data_shuffled_non_TaCo_folder
+ "audio_data_BDI___rf___True___716845___BDI.joblib"
)
# Combine model outputs to heatmap:
df_heat = pd.concat(
[
pd.DataFrame(
dict(
raw_original=model_raw.named_steps.rf.feature_importances_,
)
),
pd.DataFrame(
dict(
rem_original=model_rem.named_steps.rf.feature_importances_,
)
),
pd.DataFrame(
dict(
raw_shuffled=model_raw_shuffled.named_steps.rf.feature_importances_,
)
),
pd.DataFrame(
dict(
rem_shuffled=model_rem_shuffled.named_steps.rf.feature_importances_,
)
),
],
axis=1,
)
Preprare Feature Importance for Plotting:
df_heat = (
df_heat.set_axis(X, axis="index")
.assign(
rank_raw=lambda df: df.raw_original.rank(ascending=False, method="first"),
rank_rem=lambda df: df.rem_original.rank(ascending=False, method="first"),
# Scale Feature Importance to be easy to interpret
raw_original=lambda df: MinMaxScaler().fit_transform(df[["raw_original"]]),
rem_original=lambda df: MinMaxScaler().fit_transform(df[["rem_original"]]),
)
.pipe(lambda df: df.set_index(df.index.map(lambda x: int(x.split("__")[0]))))
)
most_imp_feat_raw_10 = df_heat.rank_raw.sort_values().iloc[:10].index.to_list()
most_imp_feat_rem_10 = df_heat.rank_rem.sort_values().iloc[:10].index.to_list()
most_important_20 = [*most_imp_feat_raw_10, *most_imp_feat_rem_10]
# Assert Non overlapping
assert len(set(most_important_20)) == 20
vmin, vmax = 0, 1
df_feat_heat_rem = df_heat.loc[most_important_20, ["rem_original", "rank_rem"]]
df_feat_heat_raw = df_heat.loc[most_important_20, ["raw_original", "rank_raw"]]
Plotting Feature Importance
fig, ax = plt.subplots()
sns.heatmap(
df_feat_heat_raw[["raw_original"]],
annot=df_feat_heat_raw[["rank_raw"]],
fmt=".5g",
vmin=vmin,
vmax=vmax,
ax=ax,
yticklabels=df_feat_heat_raw.index,
)
ax.set_title("Feature Importance Using $X$")
ax.set_ylabel("Feature Name")
ax.set_xticklabels([])
ax.set_xlabel("Feature Importance Rank")
fig.savefig("./saved_figures/feature_importance_raw.svg")
fig.savefig("./saved_figures/feature_importance_raw.png")
glue("raw_feature_importance", fig)
fig, ax = plt.subplots()
sns.heatmap(
df_feat_heat_rem[["rem_original"]],
annot=df_feat_heat_rem[["rank_rem"]],
fmt=".5g",
vmin=vmin,
vmax=vmax,
ax=ax,
yticklabels=df_feat_heat_rem.index,
)
ax.set_title("Feature Importance Using $X_{CR}$")
ax.set_ylabel("Feature Name")
ax.set_xticklabels([])
ax.set_xlabel("Feature Importance Rank")
fig.savefig("./saved_figures/feature_importance_removed.svg")
fig.savefig("./saved_figures/feature_importance_remvoed.png")
glue("rem_feature_importance", fig)
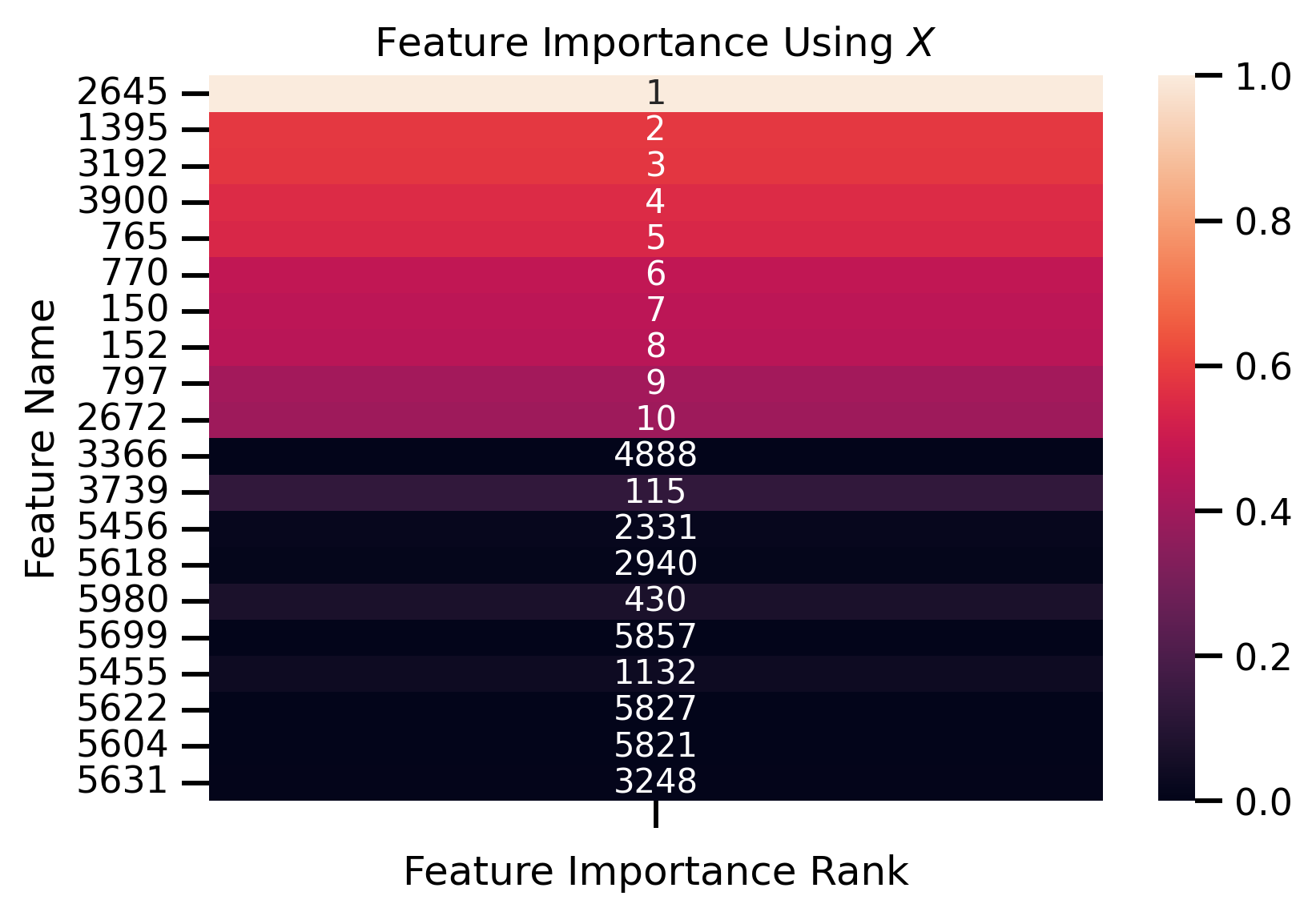
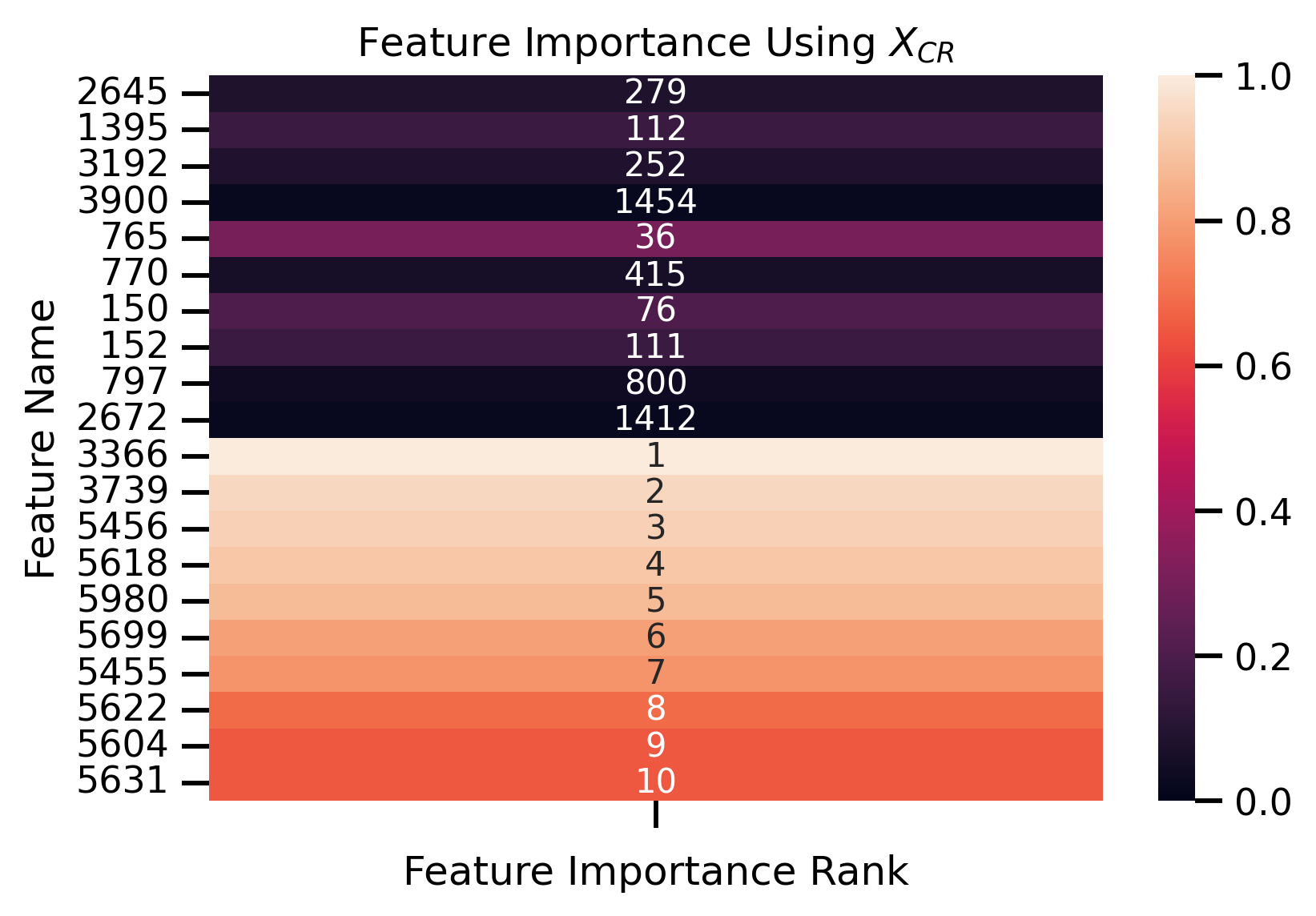
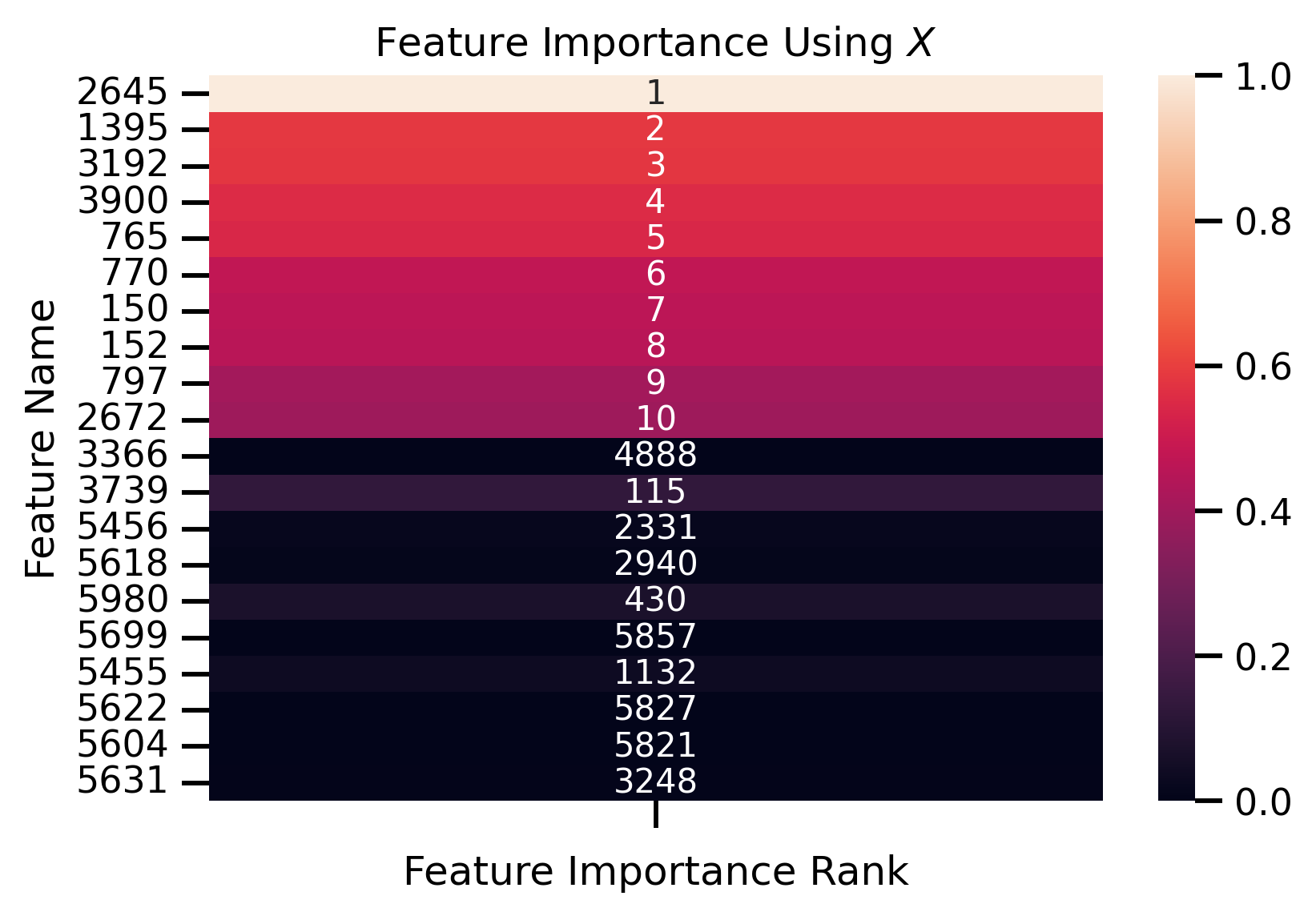
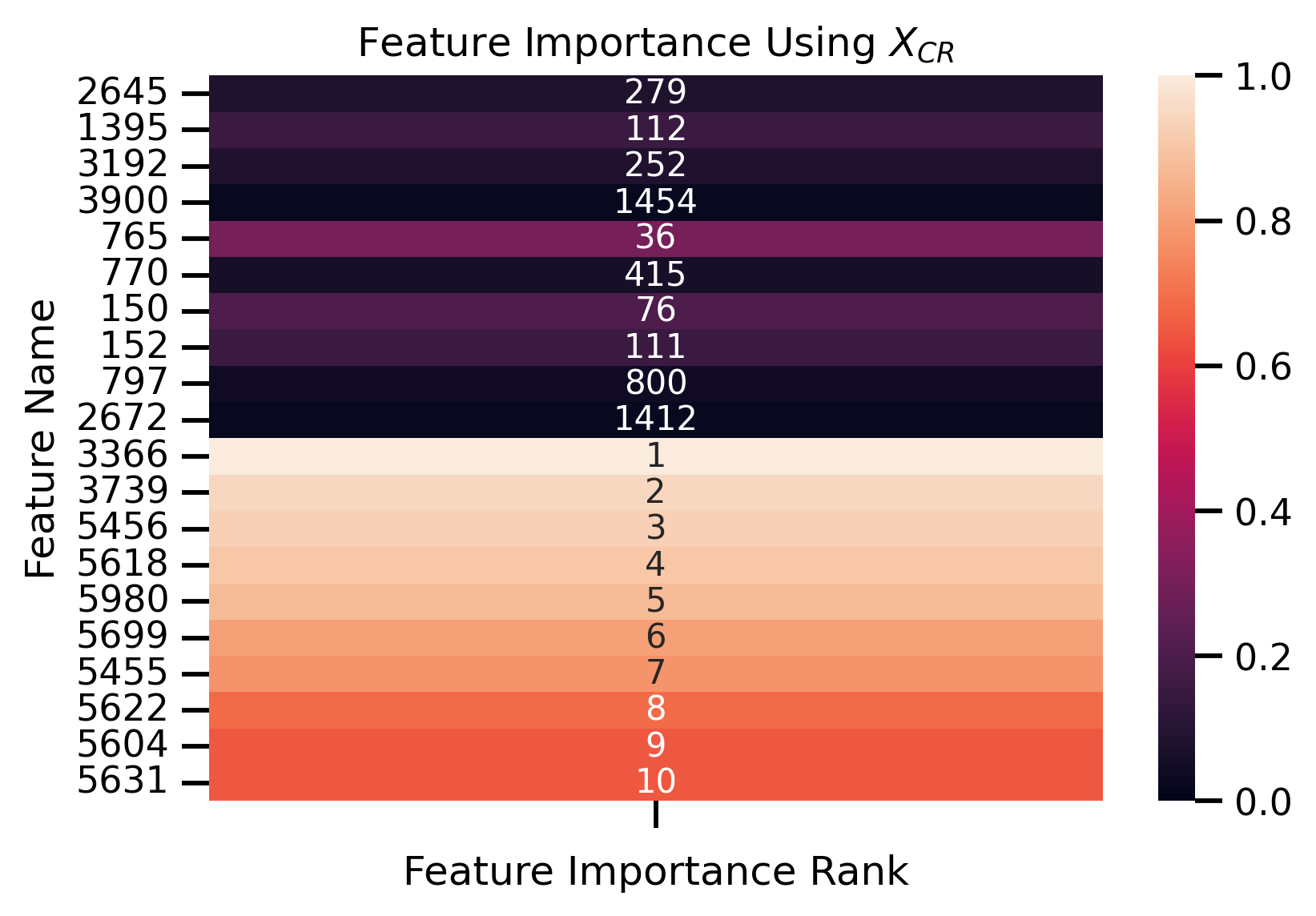
Fit new decision tree only using most important removed features:
most_imp_feat_raw = [str(i) + "__continuous" for i in most_imp_feat_raw_10[:2]]
most_imp_feat_rem = [str(i) + "__continuous" for i in most_imp_feat_rem_10[:2]]
scores_raw, model_raw = run_cross_validation(
X=most_imp_feat_rem,
y=y,
data=df_original,
model=DecisionTreeClassifier(),
preprocess_X=["zscore"],
return_estimator="final",
seed=42,
)
scores_rm, model_rm = run_cross_validation(
X=most_imp_feat_rem,
confounds=confounds,
y=y,
data=df_original,
model=DecisionTreeClassifier(),
preprocess_X=["zscore", "remove_confound"],
return_estimator="final",
seed=42,
)
print("RM:", scores_rm["test_score"].mean(), "Raw:", scores_raw["test_score"].mean())
# preprocess Features
Xcr, _ = model_rm.preprocess(df_original[most_imp_feat_rem + confounds], df_original[y])
Xraw, _ = model_raw.preprocess(df_original[most_imp_feat_rem], df_original[y])
RM: 0.7000615384615385 Raw: 0.611323076923077
Plot Raw
raw_plot = sns.jointplot(
x=most_imp_feat_rem[0],
y=most_imp_feat_rem[1],
data=Xraw.assign(**{y: df_original[y]}),
kind="scatter",
hue=y,
s=50,
)
DecisionBoundaryDisplay.from_estimator(
model_raw.named_steps.decisiontreeclassifier,
Xraw,
ax=raw_plot.ax_joint,
cmap=(LinearSegmentedColormap.from_list("MyCmapName", ["#e35d6d", "#3498eb"])),
alpha=0.5,
)
raw_plot.ax_joint.set_xlim(-1.5, 2)
raw_plot.ax_joint.set_ylim(-1, 2)
raw_plot.ax_joint.set(
xlabel="Feature: " + raw_plot.ax_joint.get_xlabel().split("_")[0],
ylabel="Feature: " + raw_plot.ax_joint.get_ylabel().split("_")[0],
)
fig_raw_decision = raw_plot.fig
fig_raw_decision.savefig("./saved_figures/decision_boundary_feature_importance_raw.svg")
fig_raw_decision.savefig("./saved_figures/decision_boundary_feature_importance_raw.png")
glue("raw_decision", fig_raw_decision)
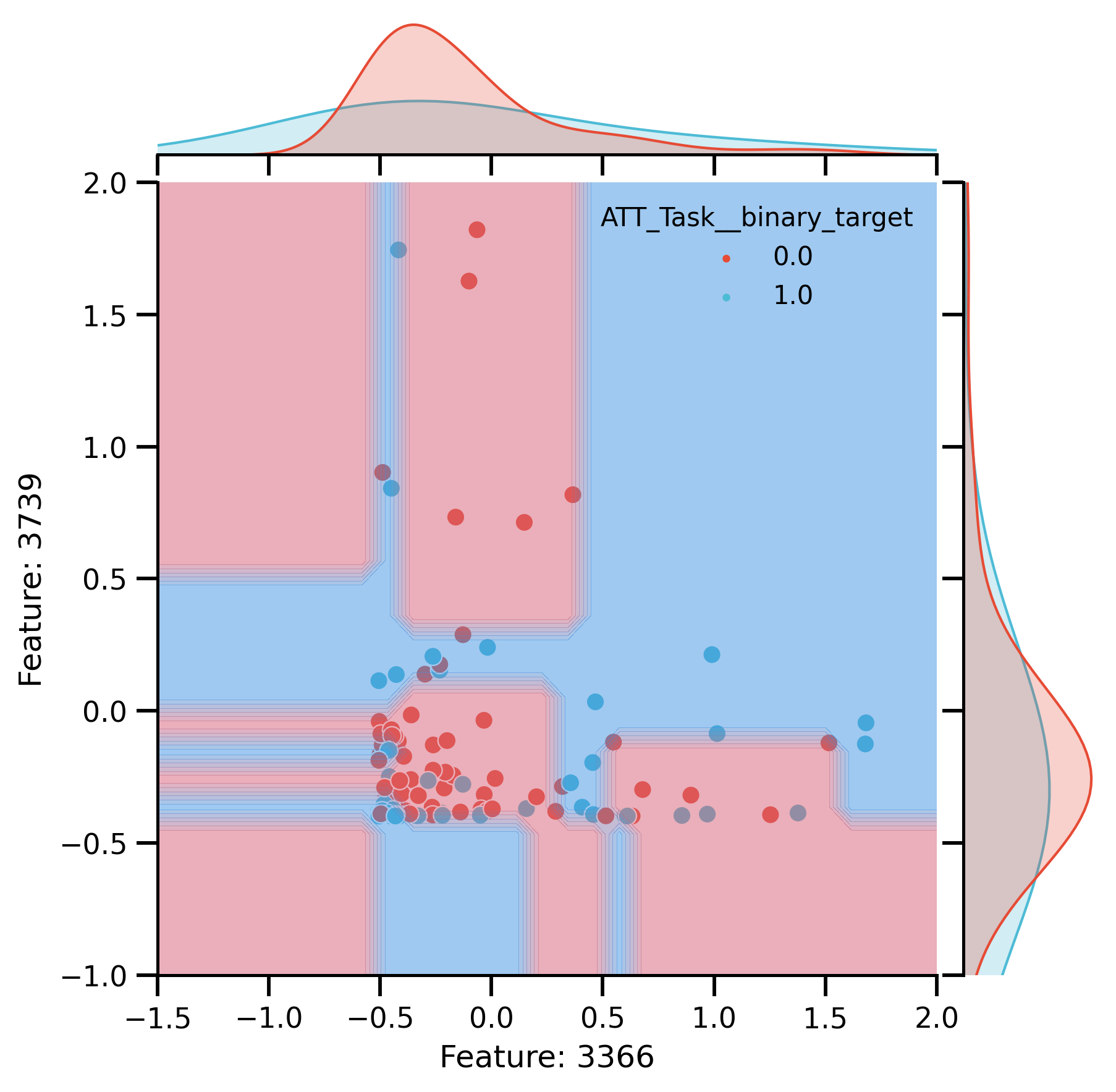
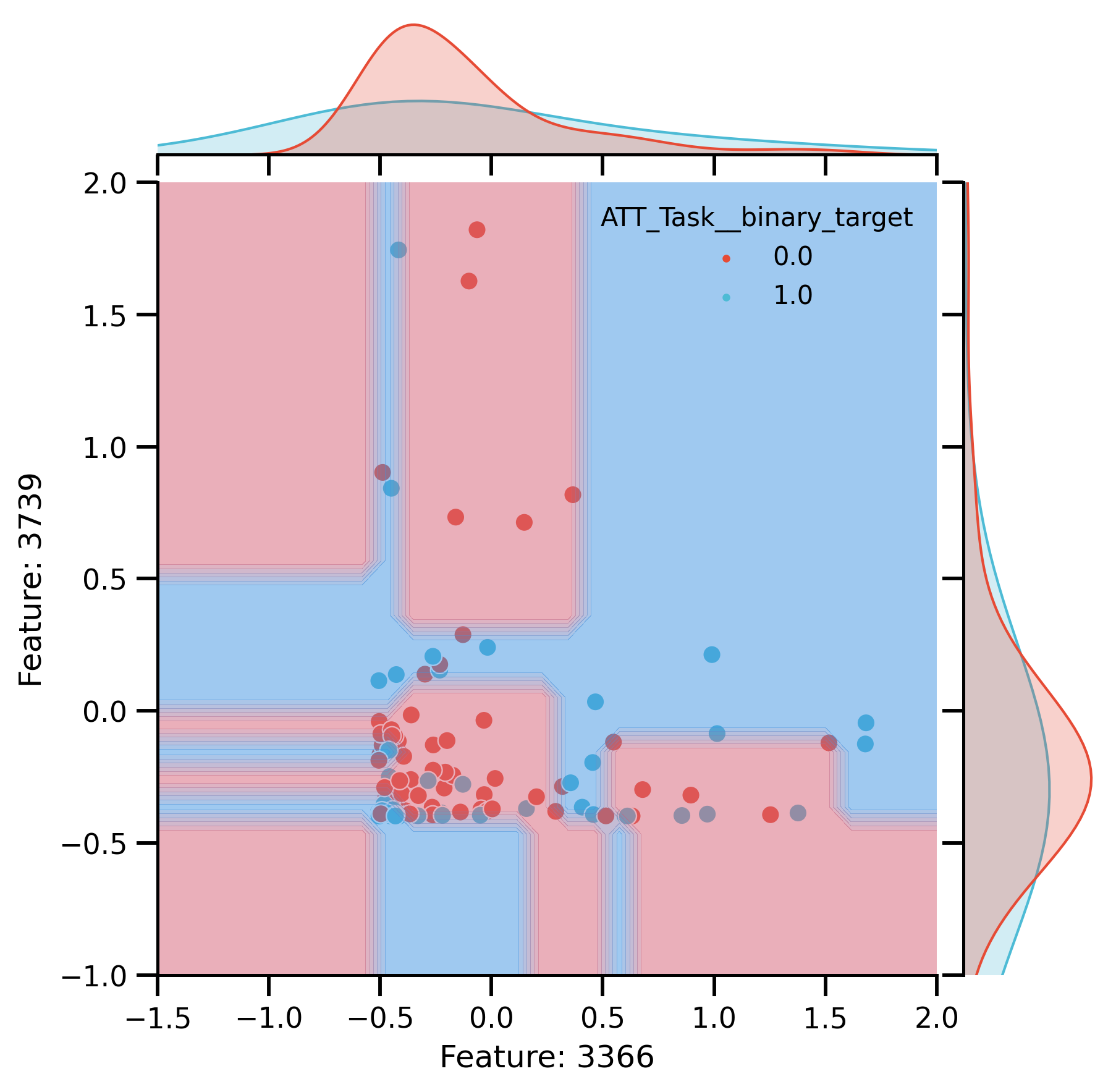
Plot after CR:
rem_plot = sns.jointplot(
x=most_imp_feat_rem[0],
y=most_imp_feat_rem[1],
data=Xcr.assign(**{y: df_original[y]}),
kind="scatter",
s=50,
hue=y,
)
rem_plot.ax_joint.set_xlim(-1.5, 2)
rem_plot.ax_joint.set_ylim(-1, 2)
DecisionBoundaryDisplay.from_estimator(
model_rm.named_steps.decisiontreeclassifier,
Xcr,
ax=rem_plot.ax_joint,
cmap=(LinearSegmentedColormap.from_list("MyCmapName", ["#e35d6d", "#3498eb"])),
alpha=0.5,
)
rem_plot.ax_joint.set(
xlabel="Feature: " + rem_plot.ax_joint.get_xlabel().split("_")[0],
ylabel="Feature: " + rem_plot.ax_joint.get_ylabel().split("_")[0],
)
fig_rem_decision = rem_plot.fig
fig_rem_decision.savefig(
"./saved_figures/decision_boundary_feature_importance_removed.svg"
)
fig_rem_decision.savefig(
"./saved_figures/decision_boundary_feature_importance_removed.png"
)
glue("rem_decision", fig_rem_decision)
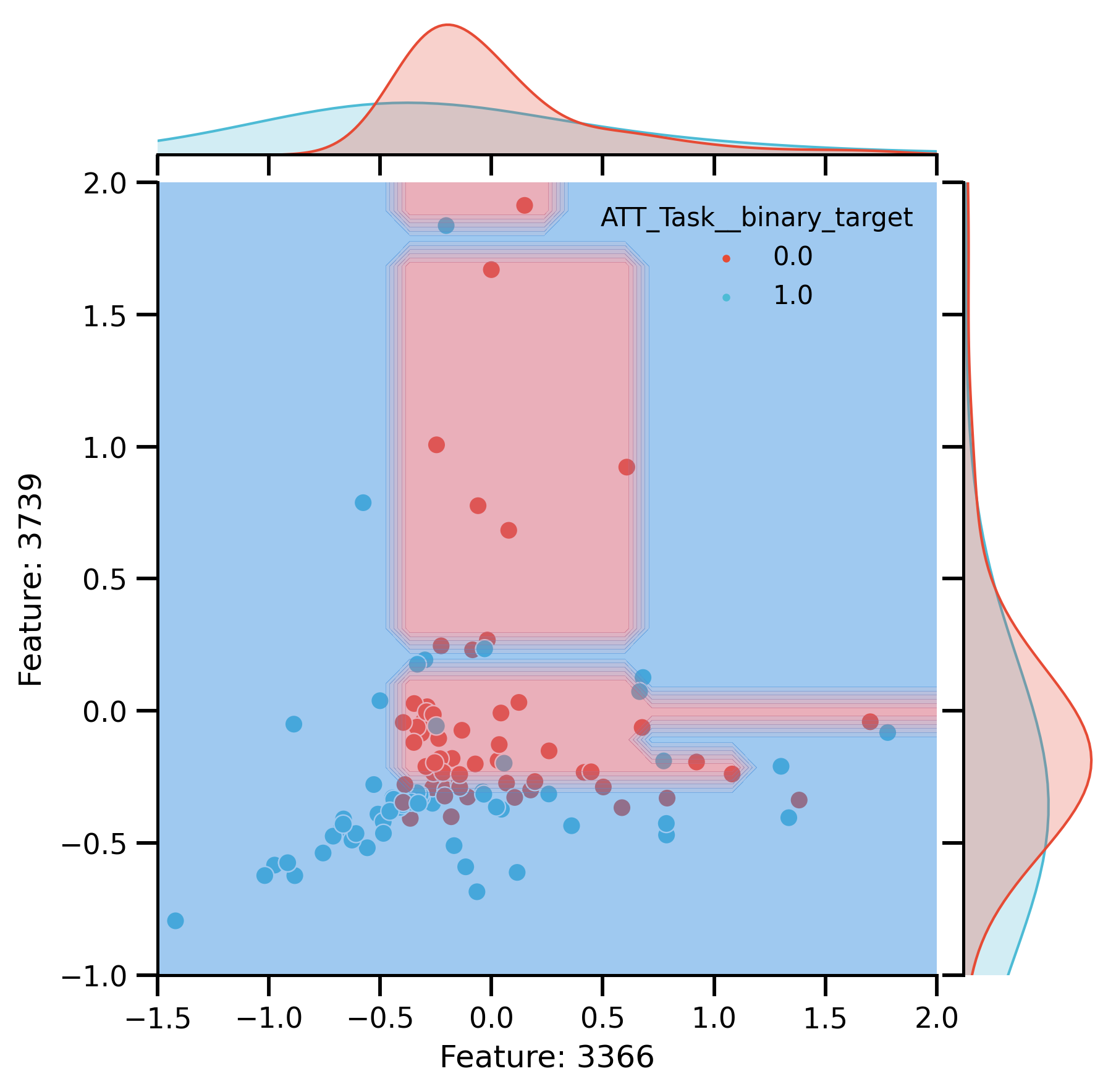
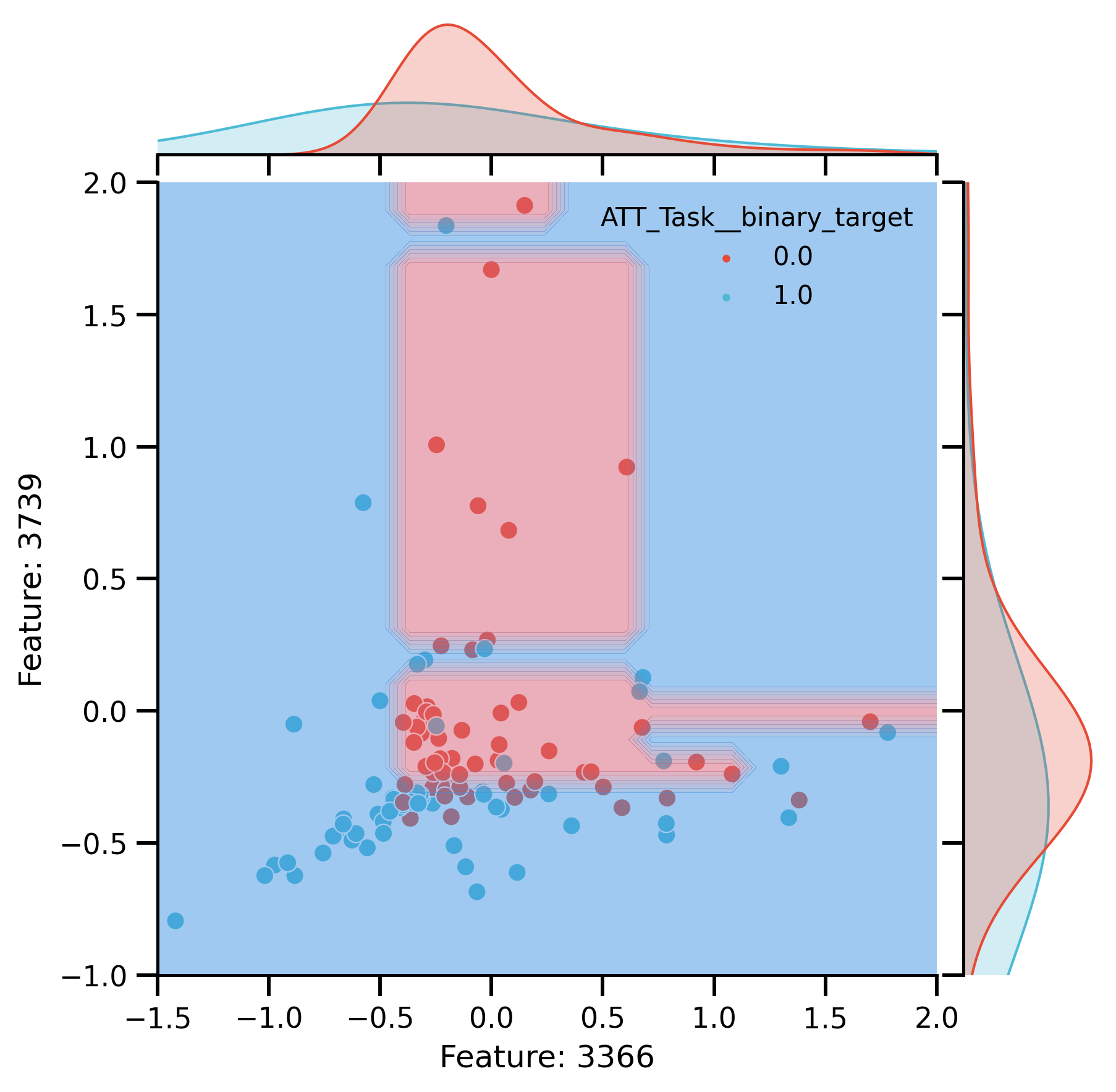
Features Importance


Decision boundaries of DT trained on 2 most important: Features.

