2c Simulation Adding more Features
2c Simulation Adding more Features¶
Prepare Imports & Paths:
import matplotlib.pyplot as plt
from scipy.stats import chi2
import numpy as np
import pandas as pd
import seaborn as sns
from julearn import run_cross_validation
from sklearn.model_selection import RepeatedKFold
import matplotlib as mpl
from leakconfound.plotting import mm_to_inch
from leakconfound.transformers import Shuffle
from sciplotlib import style
from pathlib import Path
from leakconfound.analyses.utils import save_paper_val
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
base_save_paper = "./paper_val/"
increase_features_extra_save_paper = "./paper_val/increase_features_extra/"
Path(increase_features_extra_save_paper).mkdir(parents=True, exist_ok=True)
mpl.style.use(style.get_style("nature-reviews"))
mpl.rc("xtick", labelsize=11)
mpl.rc("ytick", labelsize=11)
mpl.rc("axes", labelsize=12, titlesize=12)
mpl.rc("figure", dpi=300)
mpl.rc("figure.subplot", wspace=mm_to_inch(4), hspace=mm_to_inch(7))
mpl.rc("lines", linewidth=1)
colors = [
"#E64B35",
"#4DBBD5",
"#00A087",
"#3C5488",
"#F39B7F",
"#8491B4",
"#91D1C2FF",
"#DC0000",
"#7E6148",
"#B09C85",
]
red = colors[0]
blue = colors[1]
green = colors[2]
purple = colors[5]
Simulate Data:
np.random.seed(7329847)
confound = np.array([0] * 1000 + [1] * 1000)
target = np.random.normal(scale=0.2, size=2000) + confound
n_feat = 1
all_scores_rem = []
all_yerr_rem = []
all_scores_raw = []
all_yerr_raw = []
np.random.seed(78901234)
feat_nbr = [1, *list(range(10, 101, 10))]
for i in feat_nbr:
n_feat = i
feat = np.append(
chi2.rvs(df=3, scale=0.5, size=[1000, n_feat]),
chi2.rvs(df=4, size=[1000, n_feat]),
axis=0,
)
X = [f"feat_{ii}" for ii in range(n_feat)]
df = pd.DataFrame(feat, columns=X)
df[X] = Shuffle().fit_transform(df[X])
df["conf"] = confound
df["target"] = target
corrs = []
scores_raw = run_cross_validation(
X=X,
y="target",
data=df,
model="rf",
preprocess_X=["zscore"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
seed=293,
)
scores_rem = run_cross_validation(
X=X,
y="target",
data=df,
confounds=["conf"],
model="rf",
preprocess_X=["zscore", "remove_confound"],
preprocess_confounds="zscore",
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
seed=293,
)
scores_rem = scores_rem.groupby("repeat")["test_score"].mean()
scores_raw = scores_raw.groupby("repeat")["test_score"].mean()
all_scores_rem.append(scores_rem.mean())
all_yerr_rem.append(scores_rem.std())
all_scores_raw.append(scores_raw.mean())
all_yerr_raw.append(scores_raw.std())
save_paper_val(
increase_features_extra_save_paper,
"reasons_continuous_feat",
"raw",
f"mean_iter_{i}.txt",
scores_raw.mean(),
)
save_paper_val(
increase_features_extra_save_paper,
"reasons_continuous_feat",
"raw",
f"std_iter_{i}.txt",
scores_raw.std(),
)
save_paper_val(
increase_features_extra_save_paper,
"reasons_continuous_feat",
"removed",
f"mean_iter_{i}.txt",
scores_rem.mean(),
)
save_paper_val(
increase_features_extra_save_paper,
"reasons_continuous_feat",
"removed",
f"std_iter_{i}.txt",
scores_rem.std(),
)
/tmp/ipykernel_15745/3014949160.py:27: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
df["conf"] = confound
/tmp/ipykernel_15745/3014949160.py:28: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
df["target"] = target
df_rem = pd.DataFrame(
dict(scores=all_scores_rem, iteration=feat_nbr, yerr=all_yerr_rem)
)
df_rem["remove confounds"] = "confound removed"
df_no_rem = pd.DataFrame(
dict(scores=all_scores_raw, iteration=feat_nbr, yerr=all_yerr_raw)
)
df_no_rem["remove confounds"] = "no removal"
df_plot = pd.concat([df_rem, df_no_rem])
df_plot["confound"] = df_plot["remove confounds"].apply(
lambda x: ("removed" if x == "confound removed" else "not removed")
)
fig, ax = plt.subplots(figsize=[mm_to_inch(180), mm_to_inch(40)])
g = sns.lineplot(
x="iteration",
y="scores",
hue="confound",
hue_order=["not removed", "removed"],
data=df_plot.reset_index(),
palette=sns.color_palette([blue, red]),
ax=ax,
)
g.set(
xlabel="Number Of Features", ylabel="R²", title="χ2 Distributed Confounded Features"
)
df_plot_removed = df_plot.query('confound=="removed"')
ax.errorbar(
df_plot_removed["iteration"],
df_plot_removed["scores"],
df_plot_removed["yerr"],
fmt="none",
color=red,
)
df_plot_not_removed = df_plot.query('confound=="not removed"')
ax.errorbar(
df_plot_not_removed["iteration"],
df_plot_not_removed["scores"],
df_plot_not_removed["yerr"],
fmt="none",
color=blue,
)
ax.yaxis.set_major_locator(mpl.ticker.FixedLocator([-0.4, -0.2, 0, 0.2]))
ax.yaxis.set_major_formatter(mpl.ticker.FixedFormatter([-0.4, -0.2, 0, 0.2]))
ax.set_ylim(-0.4, 0.3)
(-0.4, 0.3)
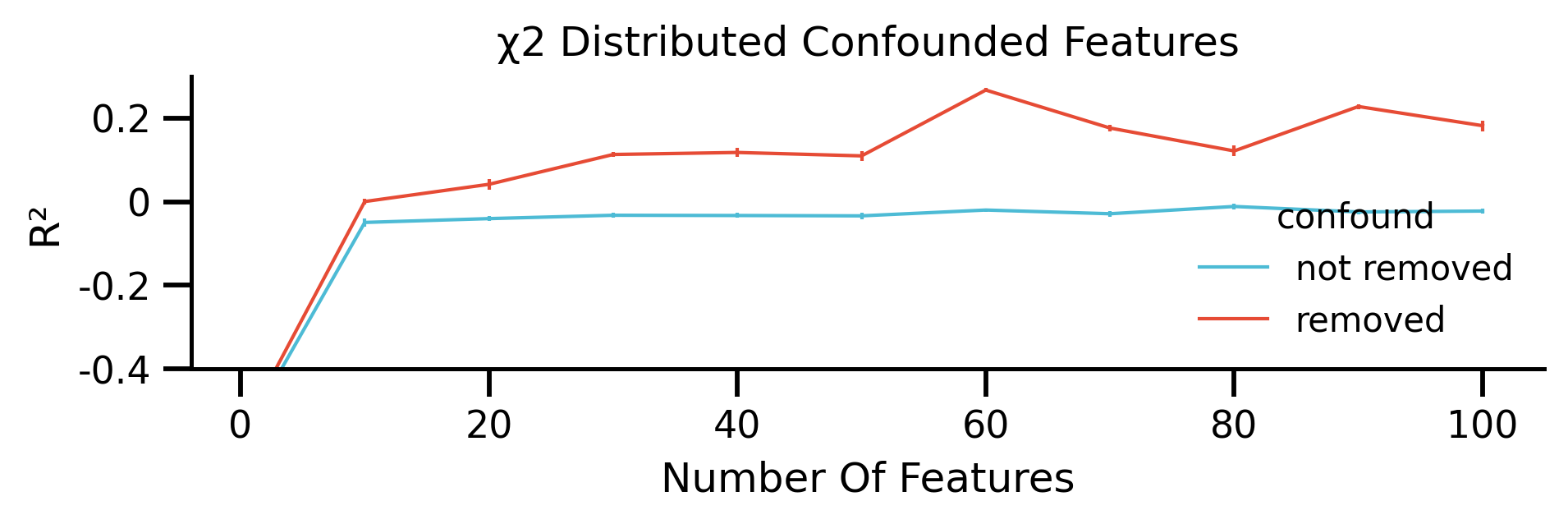
fig.savefig("./saved_figures/extra_sim_added_feat.png")
fig.savefig("./saved_figures/extra_sim_added_feat.svg")