2b Reason: Low Precision Features
Contents
2b Reason: Low Precision Features¶
Here, I will analyze how low precision in the features can influence confound-leakage. The lowest precision of a feature would be binary 0 and 1, but later I will also look at precisions, where steps are lower than 1.
Configure imports and path:
from matplotlib import ticker
from functools import partial
from copy import copy
import matplotlib as mpl
import numpy as np
import joblib
import pandas as pd
from julearn import run_cross_validation
from sklearn.model_selection import RepeatedKFold, KFold, RepeatedStratifiedKFold
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
import seaborn as sns
import matplotlib.pyplot as plt
from dtreeviz.trees import dtreeviz
from leakconfound.plotting import mm_to_inch
from leakconfound.analyses.utils import save_paper_val
from sciplotlib import style
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
np.random.seed(6252006)
base_save_paper = "./paper_val/"
base_dir = "../../"
colors = [
"#E64B35",
"#4DBBD5",
"#00A087",
"#3C5488",
"#F39B7F",
"#8491B4",
"#91D1C2FF",
"#DC0000",
"#7E6148",
"#B09C85",
]
red = colors[0]
blue = colors[1]
green = colors[2]
purple = colors[5]
mpl.style.use(style.get_style("nature-reviews"))
mpl.rc("xtick", labelsize=11)
mpl.rc("ytick", labelsize=11)
mpl.rc("axes", labelsize=12, titlesize=12)
mpl.rc("figure", dpi=300)
mpl.rc("figure.subplot", wspace=mm_to_inch(4), hspace=mm_to_inch(7))
mpl.rc("lines", linewidth=1)
Define some helper functions to be able to sample from specific distributions.
def convert_precision(original_nbr, step=0.2):
if original_nbr % step > step / 2:
res = step * (original_nbr // step + 1)
else:
res = step * (original_nbr // step)
return res
def normal_stepped(size, step=0.2):
df_normal = pd.DataFrame(np.random.normal(size=size), columns=["feat_1"])
return df_normal.assign(
feat_2=lambda df: df["feat_1"].map(partial(convert_precision, step=step))
)
Starting with Binary Feature¶
target = np.random.normal(size=1_000)
feat = np.random.choice([0, 1], replace=True, size=1_000)
df = pd.DataFrame(dict(feat_1=feat, conf=target, target=target))
scores_1_no_rem, est_no_removal = run_cross_validation(
X=["feat_1"],
y="target",
data=df,
model=DecisionTreeRegressor(),
preprocess_X=["zscore"],
preprocess_confounds=["zscore"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
return_estimator="final",
)
scores_1_no_rem = scores_1_no_rem.groupby("repeat").mean().test_score
scores_1, est_removed = run_cross_validation(
X=["feat_1"],
confounds=["conf"],
y="target",
data=df,
model=DecisionTreeRegressor(),
preprocess_X=["zscore", "remove_confound"],
preprocess_confounds=["zscore"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
return_estimator="final",
)
scores_1 = scores_1.groupby("repeat").mean().test_score
print(
"Scores random binary no removal feature : M = "
f"{scores_1_no_rem.mean():.4f}, sd = {scores_1_no_rem.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"mean_random_binary.txt",
scores_1_no_rem.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"std_random_binary.txt",
scores_1_no_rem.std(),
)
print(
"Scores random binary confound removed feature : M = "
f"{scores_1.mean():.4f}, sd = {scores_1.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_random_binary.txt",
scores_1.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_random_binary.txt",
scores_1.std(),
)
Scores random binary no removal feature : M = -0.0068, sd = 0.0034
Scores random binary confound removed feature : M = 0.9985, sd = 0.0003
Now let us have a closer look at what is going on:
target = np.random.normal(size=1_000)
feat = np.random.choice([0, 1], replace=True, size=1_000)
df_test = pd.DataFrame(dict(feat_1=feat, conf=target, target=target))
pred_rem = est_removed.predict(df_test[["feat_1", "conf"]])
pred_no_rem = est_no_removal.predict(df_test[["feat_1"]])
X_prep_rem, _ = est_removed.preprocess(df_test.drop(columns=["target"]), df_test.target)
X_prep_no_rem, _ = est_no_removal.preprocess(
df_test.drop(columns=["target", "conf"]), df_test.target
)
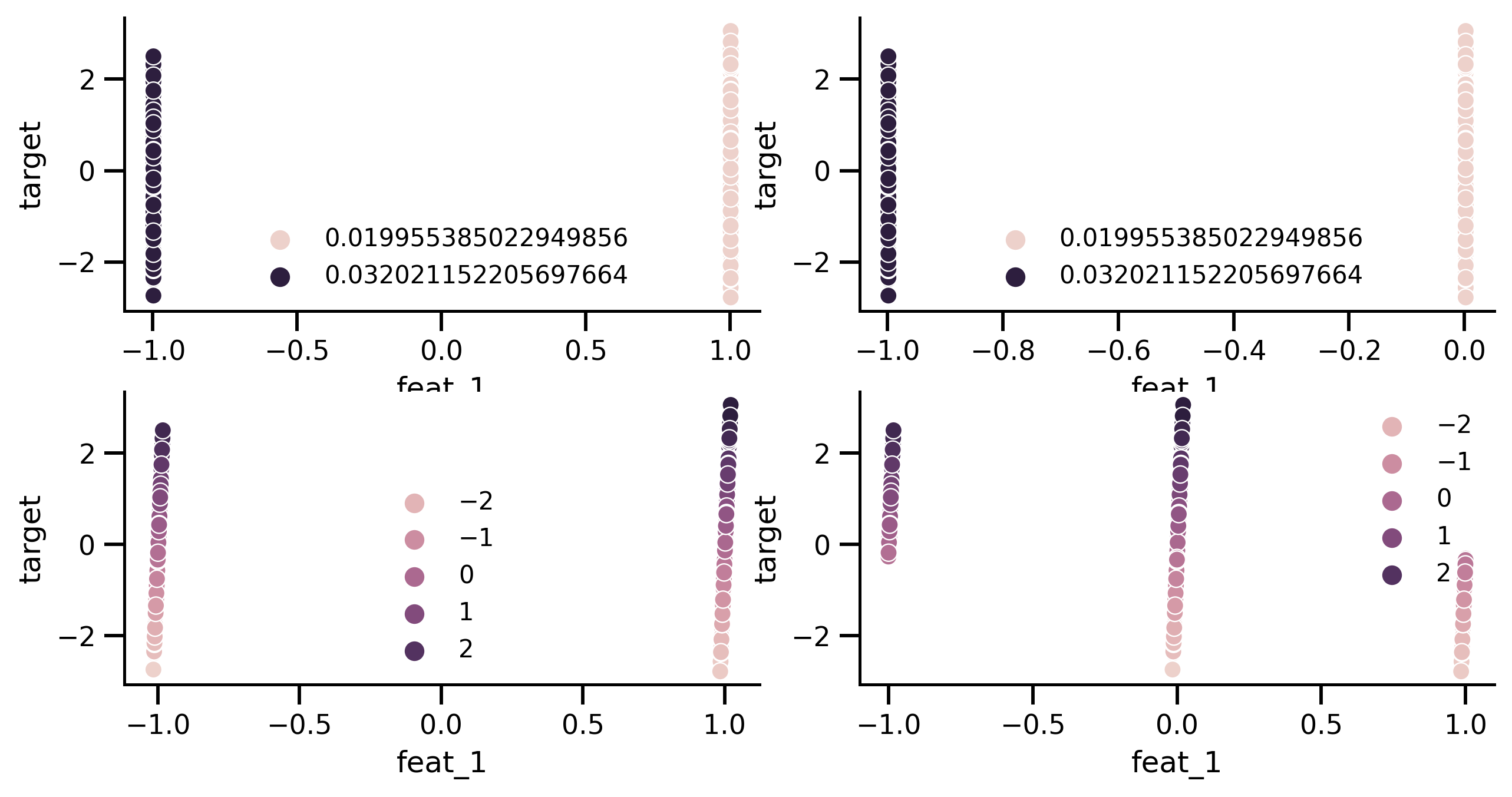
fig, axes = plt.subplots(2, 2, figsize=[10, 5])
sns.scatterplot(
x=X_prep_no_rem["feat_1"], y=df_test.target, ax=axes[0][0], hue=pred_no_rem
)
sns.scatterplot(
x=X_prep_no_rem["feat_1"].apply(lambda x: x - int(x)),
y=df_test.target,
ax=axes[0][1],
hue=pred_no_rem,
)
sns.scatterplot(x=X_prep_rem["feat_1"], y=df_test.target, ax=axes[1][0], hue=pred_rem)
sns.scatterplot(
x=X_prep_rem["feat_1"].apply(lambda x: x - int(x)),
y=df_test.target,
ax=axes[1][1],
hue=pred_rem,
)
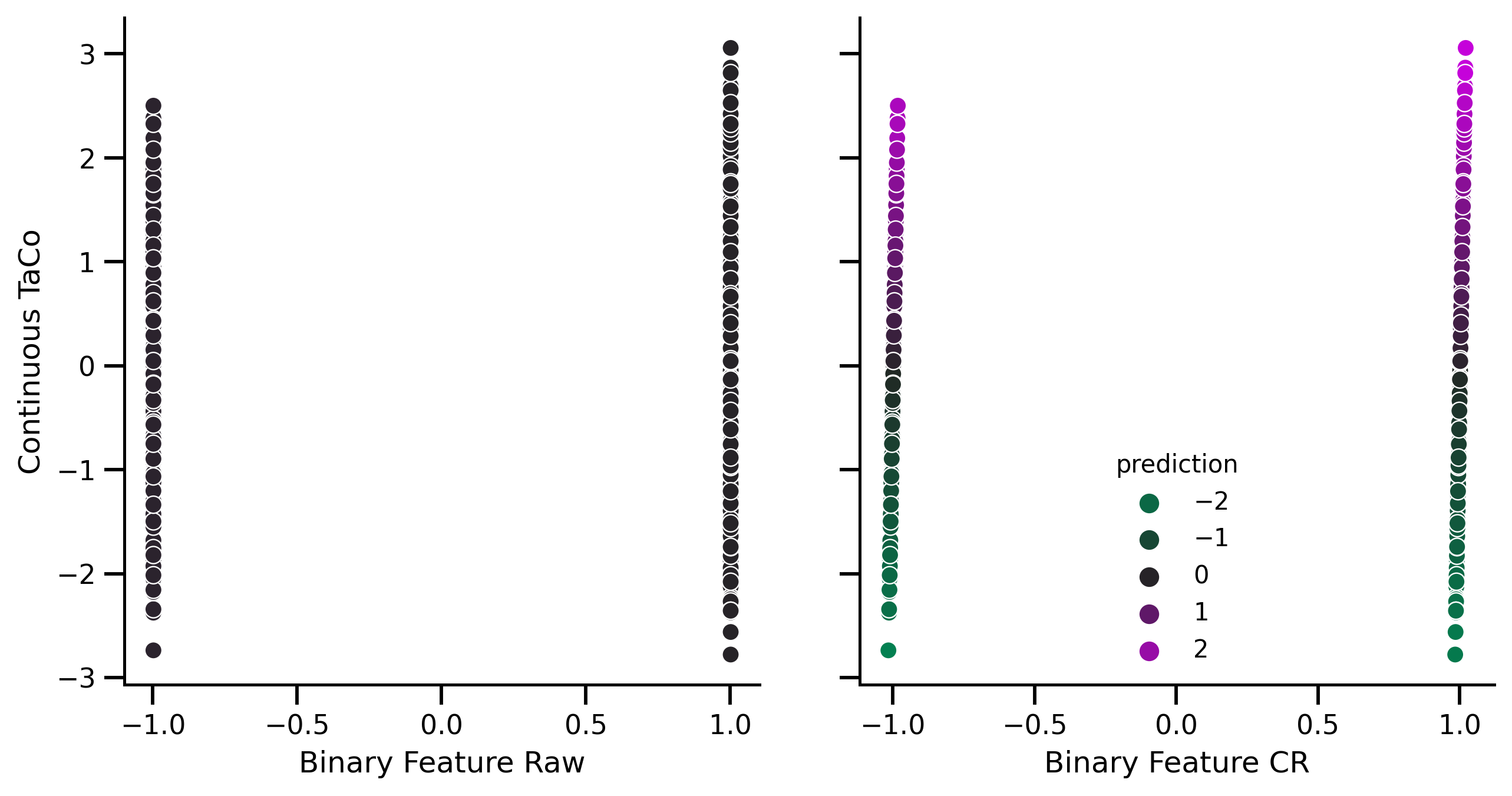
fig, axes = plt.subplots(1, 2, figsize=[10, 5])
palette = sns.diverging_palette(
145,
300,
s=100,
as_cmap=True,
center="dark",
)
normalize = mpl.colors.Normalize(vmin=-3, vmax=3)
sns.scatterplot(
x=X_prep_no_rem["feat_1"],
y=df_test.target,
ax=axes[0],
palette=palette,
hue=pred_no_rem,
hue_norm=normalize,
)
sns.scatterplot(
x=X_prep_rem["feat_1"],
y=df_test.target,
ax=axes[1],
palette=palette,
hue=pred_rem,
hue_norm=normalize,
)
axes[0].get_legend().set_visible(False)
axes[1].get_legend().set_title("prediction")
axes[0].set_xlabel("Binary Feature Raw")
axes[0].set_ylabel("Continuous TaCo")
axes[1].set_xlabel("Binary Feature CR")
axes[1].set_yticklabels([])
axes[1].set_ylabel("")
fig.savefig("./saved_figures/low_precision_sim.svg")
Investigating Similar Effect for Continuous Values Rounded to a Precision¶
step = 0.1
target = np.random.normal(size=1_000)
df = normal_stepped(size=1_000, step=step)
df["target"] = target
df["conf"] = target
scores_1 = (
run_cross_validation(
X=["feat_1"],
y="target",
data=df,
confounds=["conf"],
model=DecisionTreeRegressor(),
preprocess_X=["zscore", "remove_confound"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
)
.groupby("repeat")
.mean()
.test_score
)
scores_1_no_cr = (
run_cross_validation(
X=["feat_1"],
y="target",
data=df,
model=DecisionTreeRegressor(),
preprocess_X=["zscore"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
)
.groupby("repeat")
.mean()
.test_score
)
scores_2, est_removed = run_cross_validation(
X=["feat_2"],
y="target",
data=df,
confounds=["conf"],
model=DecisionTreeRegressor(),
preprocess_X=["zscore", "remove_confound"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
return_estimator="final",
)
scores_2 = scores_2.groupby("repeat").mean().test_score
scores_3, est_no_removal = run_cross_validation(
X=["feat_2"],
y="target",
data=df,
model=DecisionTreeRegressor(),
preprocess_X=["zscore"],
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
return_estimator="final",
)
scores_3 = scores_3.groupby("repeat").mean().test_score
print(
"Scores Random confound no removal continuous Feature: M = "
f"{scores_1_no_cr.mean():.4f}, sd = {scores_1_no_cr.std():.4f}"
)
print(
"Scores Random confound removed continuous Feature: M = "
f"{scores_1.mean():.4f}, sd = {scores_1.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"mean_random_continious_cont_target_not_rounded.txt",
scores_1_no_cr.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"std_random_continious_cont_target_not_rounded.txt",
scores_1_no_cr.std(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_random_continious_cont_target_not_rounded.txt",
scores_1.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_random_continious_cont_target_not_rounded.txt",
scores_1.std(),
)
print(
"Scores Random confound removed continuous Feature with"
f"precision of {step} : M = "
f"{scores_2.mean():.4f}, sd = {scores_2.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_random_continious_cont_target_prec_rounded.txt",
scores_2.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_random_continious_cont_target_prec_rounded.txt",
scores_2.std(),
)
print(
"Scores Random no removal continuous Feature with "
f"precision of {step} : M = "
f"{scores_3.mean():.4f}, sd = {scores_3.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"mean_random_continious_cont_target_prec_rounded.txt",
scores_3.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"std_random_continious_cont_target_prec_rounded.txt",
scores_3.std(),
)
Scores Random confound no removal continuous Feature: M = -1.0989, sd = 0.0611
Scores Random confound removed continuous Feature: M = -1.0280, sd = 0.0717
Scores Random confound removed continuous Feature withprecision of 0.1 : M = 0.7041, sd = 0.1646
Scores Random no removal continuous Feature with precision of 0.1 : M = -0.0845, sd = 0.0126
target_test = np.random.normal(size=1_000)
df_test = normal_stepped(size=1_000, step=step)
df_test["target"] = target_test
df_test["conf"] = target_test
pred_rem = est_removed.predict(df_test[["feat_2", "conf"]])
pred_no_rem = est_no_removal.predict(df_test[["feat_2"]])
X_prep_rem, _ = est_removed.preprocess(
df_test.drop(columns=["target", "feat_1"]), df_test.target
)
X_prep_no_rem, _ = est_no_removal.preprocess(
df_test.drop(columns=["target", "feat_1", "conf"]), df_test.target
)
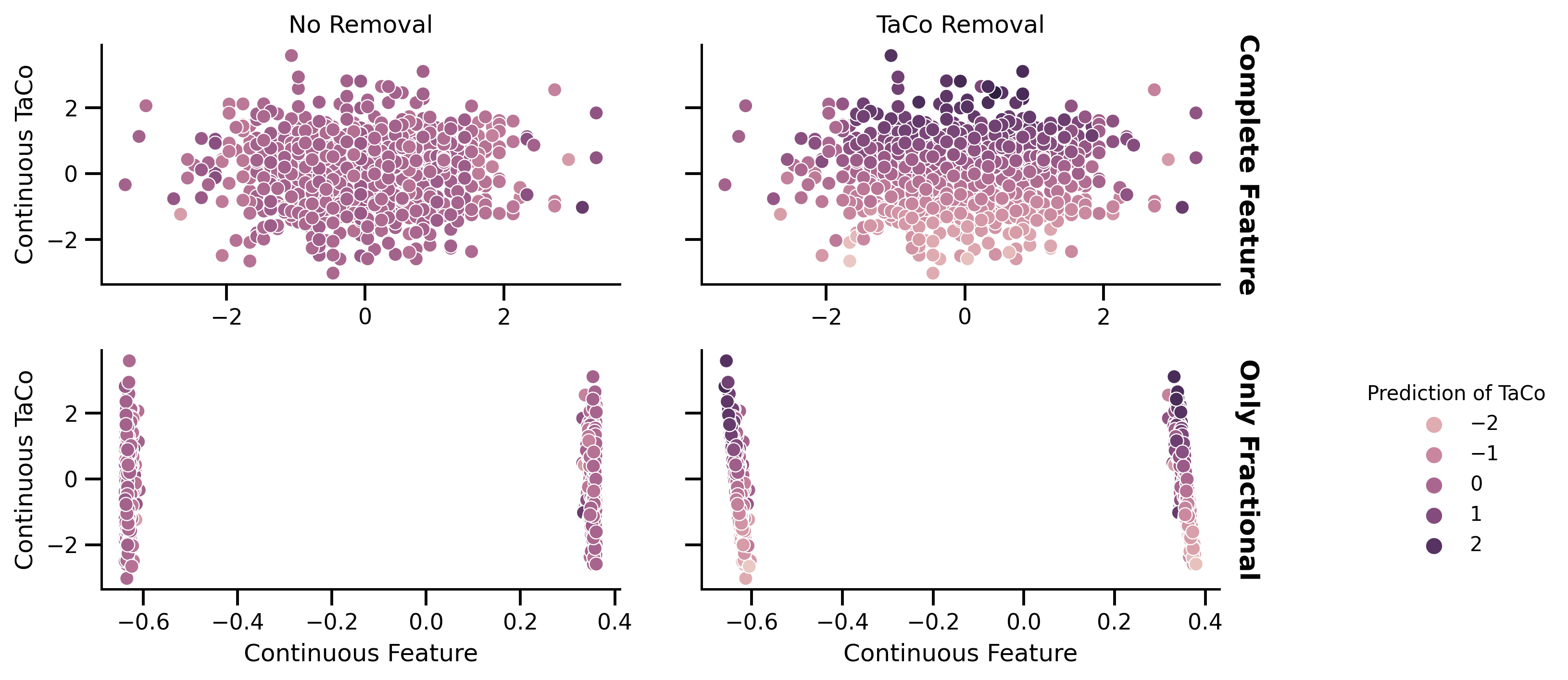
fig, axes = plt.subplots(2, 2, figsize=[10, 5])
normalize = mpl.colors.Normalize(vmin=-3, vmax=3)
sns.scatterplot(
x=X_prep_no_rem["feat_2"],
y=df_test.target,
ax=axes[0][0],
hue=pred_no_rem,
hue_norm=normalize,
)
sns.scatterplot(
x=X_prep_no_rem["feat_2"].apply(lambda x: (x * 10 - int(x * 10))),
y=df_test.target,
ax=axes[1][0],
hue=pred_no_rem,
hue_norm=normalize,
)
sns.scatterplot(
x=X_prep_rem["feat_2"],
y=df_test.target,
ax=axes[0][1],
hue=pred_rem,
hue_norm=normalize,
)
g = sns.scatterplot(
x=X_prep_rem["feat_2"].apply(lambda x: (x * 10 - int(x * 10))),
y=df_test.target,
ax=axes[1][1],
hue=pred_rem,
hue_norm=normalize,
)
axes[0][0].get_legend().set_visible(False)
axes[0][1].get_legend().set_visible(False)
axes[1][0].get_legend().set_visible(False)
g.legend(
loc="center left", bbox_to_anchor=(1.25, 0.5), ncol=1, title="Prediction of TaCo"
)
axes[0][0].set_title("No Removal")
axes[0][1].set_title("TaCo Removal")
axes[1][0].set_xlabel("Continuous Feature")
axes[1][1].set_xlabel("Continuous Feature")
axes[0][0].set_xlabel("")
axes[0][1].set_xlabel("")
axes[0][0].set_ylabel("Continuous TaCo")
axes[1][0].set_ylabel("Continuous TaCo")
axes[0][1].set_ylabel("")
axes[1][1].set_ylabel("")
axes[0][1].set_yticklabels([])
axes[1][1].set_yticklabels([])
axR0 = axes[0][1].twinx()
axR0.set_yticks([])
axR0.set_ylabel(
"Complete Feature", rotation=270, labelpad=20, fontsize=13, weight="bold"
)
axR1 = axes[1][1].twinx()
axR1.set_yticks([])
axR1.set_ylabel(
"Only Fractional", rotation=270, labelpad=20, fontsize=13, weight="bold"
)
fig.savefig("./saved_figures/low_precision_cont_sim.svg")
same but now with a binary target:
target = np.random.choice([0, 1], size=1_000)
df = normal_stepped(size=1_000, step=step)
df["target"] = target
df["conf"] = target
scores_1 = run_cross_validation(
X=["feat_1"],
y="target",
data=df,
confounds=["conf"],
model=DecisionTreeClassifier(),
preprocess_X=["zscore", "remove_confound"],
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
)
scores_1 = scores_1.groupby("repeat").mean().test_score
scores_2, est_removed = run_cross_validation(
X=["feat_2"],
y="target",
data=df,
confounds=["conf"],
model=DecisionTreeClassifier(),
preprocess_X=["zscore", "remove_confound"],
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
return_estimator="final",
)
scores_2 = scores_2.groupby("repeat").mean().test_score
scores_3, est_no_removal = run_cross_validation(
X=["feat_2"],
y="target",
data=df,
model=DecisionTreeClassifier(),
preprocess_X=["zscore"],
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
return_estimator="final",
)
scores_3 = scores_3.groupby("repeat").mean().test_score
print(
"Scores Random confound removed continuous Feature Binary Target: M = "
f"{scores_1.mean():.4f}, sd = {scores_1.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_random_continious_bin_target_prec_1.txt",
scores_1.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_random_continious_bin_target_prec_1.txt",
scores_1.std(),
)
print(
"Scores Random confound removed continuous Feature with"
f"precision of {step} Binary Target : M = "
f"{scores_2.mean():.4f}, sd = {scores_2.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_random_continious_bin_target_prec_2.txt",
scores_2.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_random_continious_bin_target_prec_2.txt",
scores_2.std(),
)
print(
"Scores Random no removal continuous Feature with "
f"precision of {step} Binary Target : M = "
f"{scores_3.mean():.4f}, sd = {scores_3.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_random_continious_bin_target_prec_3.txt",
scores_3.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_random_continious_bin_target_prec_3.txt",
scores_3.std(),
)
Scores Random confound removed continuous Feature Binary Target: M = 0.4929, sd = 0.0113
Scores Random confound removed continuous Feature withprecision of 0.1 Binary Target : M = 0.9836, sd = 0.0026
Scores Random no removal continuous Feature with precision of 0.1 Binary Target : M = 0.5265, sd = 0.0089
Now to a real dataset example from UCI “Heart” dataset using a decision tree. First, some info about the fitted model:
case = "heart___decisiontree___False___716845___num"
model_no_rm = joblib.load(
f"{base_dir}results/basic_TaCo/uci_datasets/models/{case}.joblib"
)
df_performance_no_rm = pd.read_csv(
f"{base_dir}results/basic_TaCo/uci_datasets/{case}.csv"
)
print(
f"Performance No removal M = {df_performance_no_rm.test_roc_auc.mean():.2f},"
f"sd={df_performance_no_rm.test_roc_auc.std(): .2f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"mean_heart.txt",
df_performance_no_rm.test_roc_auc.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"std_heart.txt",
df_performance_no_rm.test_roc_auc.std(),
)
print(model_no_rm.named_steps.decisiontreeclassifier.feature_importances_)
print(
f"Not removed has {model_no_rm.named_steps.decisiontreeclassifier.tree_.node_count} nodes"
)
case = "heart___decisiontree___True___716845___num"
model_rm = joblib.load(
f"{base_dir}results/basic_TaCo/uci_datasets/models/{case}.joblib"
)
print(model_rm.named_steps.decisiontreeclassifier.feature_importances_)
print(
f"removed has {model_rm.named_steps.decisiontreeclassifier.tree_.node_count} nodes"
)
df_performance_rm = pd.read_csv(f"{base_dir}results/basic_TaCo/uci_datasets/{case}.csv")
print(
f"Performance Removal M = {df_performance_rm.test_roc_auc.mean():.2f},"
f"sd={df_performance_rm.test_roc_auc.std(): .2f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_heart.txt",
df_performance_rm.test_roc_auc.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_heart.txt",
df_performance_rm.test_roc_auc.std(),
)
df = pd.read_csv(f'{base_dir}data/uci_datasets/{case.split("___")[0]}.csv')
y = "num__binary_target"
Performance No removal M = 0.74,sd= 0.06
[0.18565167 0.0558061 0.09962636 0.11968048 0.04075426 0.
0.00973236 0.00985401 0.01487661 0. 0.10911591 0.
0. 0. 0.01448795 0.05085158 0.01216545 0.27739726
0. 0. ]
Not removed has 115 nodes
[0. 0. 0. 0. 0. 0.04285714
0. 0. 0. 0. 0. 0.
0.95714286 0. 0. 0. 0. 0.
0. 0. ]
removed has 5 nodes
Performance Removal M = 0.99,sd= 0.01
Preparing data to visualize parts of the DT:
def correct_dummy_name(dummy_column):
"""names will be name__categorical_1, but should be name_1__categorical
so thate __categorical as type is still valid
"""
if dummy_column[-1].isdigit():
name, column_type = dummy_column.split("__")
column_type, number = column_type.split("_")
return f"{name}_{number}__{column_type}"
return dummy_column
confounds = ["num__categorical_confound__:type:__confound"]
df_rm = df.copy().assign(
**{"num__categorical_confound__:type:__confound": df.num__binary_target}
)
categorical = [
col
for col in df_rm.copy().columns.to_list()
if (col.endswith("categorical") or col.endswith("nominal"))
]
categorical_non_bi = [
cat for cat in categorical if df_rm[cat].copy().unique().shape[0] > 2
]
df_ohe = pd.get_dummies(df_rm[categorical], columns=categorical_non_bi)
df_rm.drop(columns=categorical, inplace=True)
categorical = [correct_dummy_name(col) for col in df_ohe.columns.to_list()]
df_rm[categorical] = df_ohe
X = [
col
for col in df_rm.columns.to_list()
if ("categorical" in col and "confound" not in col) or col.endswith("continuous")
]
df_removed, _ = model_rm.preprocess(df_rm[X + confounds].copy(), df_rm[y].copy())
X_names = copy(X)
X_names[4] = "Fasting Blood Sugar"
X_names[11] = "Resting Electrocardiographic Results (0 vs. Rest)"
dt_colors = [
None, # 0 classes
None, # 1 class
[green, purple], # 2 classes
]
viz = dtreeviz(
model_rm["decisiontreeclassifier"],
df_removed,
df[y],
target_name="Heart Disease",
feature_names=X_names + confounds,
histtype="strip",
scale=2,
colors={"classes": dt_colors},
class_names={0: "not diagnosed", 1: "diagnosed"},
)
viz.save("./saved_figures/dt_heart.svg")
findfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.
Plot Nodes as Swarm Plots:
def plot_discont(X, hue, data, left_lim, right_lim, colors=None, title=""):
df_left = data.query(f"{X} < {left_lim[1]}").copy()
df_right = data.query(f"{X} > {right_lim[0]}").copy()
assert len(df_left) + len(df_right) == len(data)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=[10, 5], sharey=True)
fig.subplots_adjust(wspace=0.05)
fig.suptitle(title)
df_right["ccount"] = df_right.groupby(y).cumcount()
y_value_counts_right = df_right[y].value_counts()
df_right["plot_y"] = df_right.apply(
lambda row: row["ccount"] / y_value_counts_right[row[y]], axis=1
)
df_left["ccount"] = df_left.groupby(y).cumcount()
y_value_counts_left = df_left[y].value_counts()
df_left["plot_y"] = df_left.apply(
lambda row: row["ccount"] / y_value_counts_left[row[y]], axis=1
)
palette_left = sns.color_palette(np.array(colors)[y_value_counts_left.index.values])
palette_right = sns.color_palette(
np.array(colors)[y_value_counts_right.index.values]
)
sns.scatterplot(
x=X, y="plot_y", hue=hue, data=df_left, ax=ax1, palette=palette_left
)
sns.scatterplot(
x=X, y="plot_y", hue=hue, data=df_right, ax=ax2, palette=palette_right
)
ax1.set_xlim(left_lim[0], left_lim[1])
ax1.get_legend().set_visible(False)
ax1.spines.right.set_visible(False)
ax1.set_yticklabels([])
ax1.set_ylabel("")
ax1.tick_params(left=False)
# ax2
ax2.spines.left.set_visible(False)
ax2.yaxis.tick_left()
ax2.tick_params(left=False)
ax2.set_xlim(right_lim[0], right_lim[1])
d = 1.5 # proportion of vertical to horizontal extent of the slanted line
kwargs = dict(
marker=[(-1, -d), (1, d)],
markersize=12,
linestyle="none",
color="k",
mec="k",
mew=1,
clip_on=False,
)
ax1.plot([1], [0], transform=ax1.transAxes, **kwargs)
ax2.plot([0], [0], transform=ax2.transAxes, **kwargs)
# titles
ax1.set_xlabel("")
ax2.set_xlabel("")
return fig, (ax1, ax2)
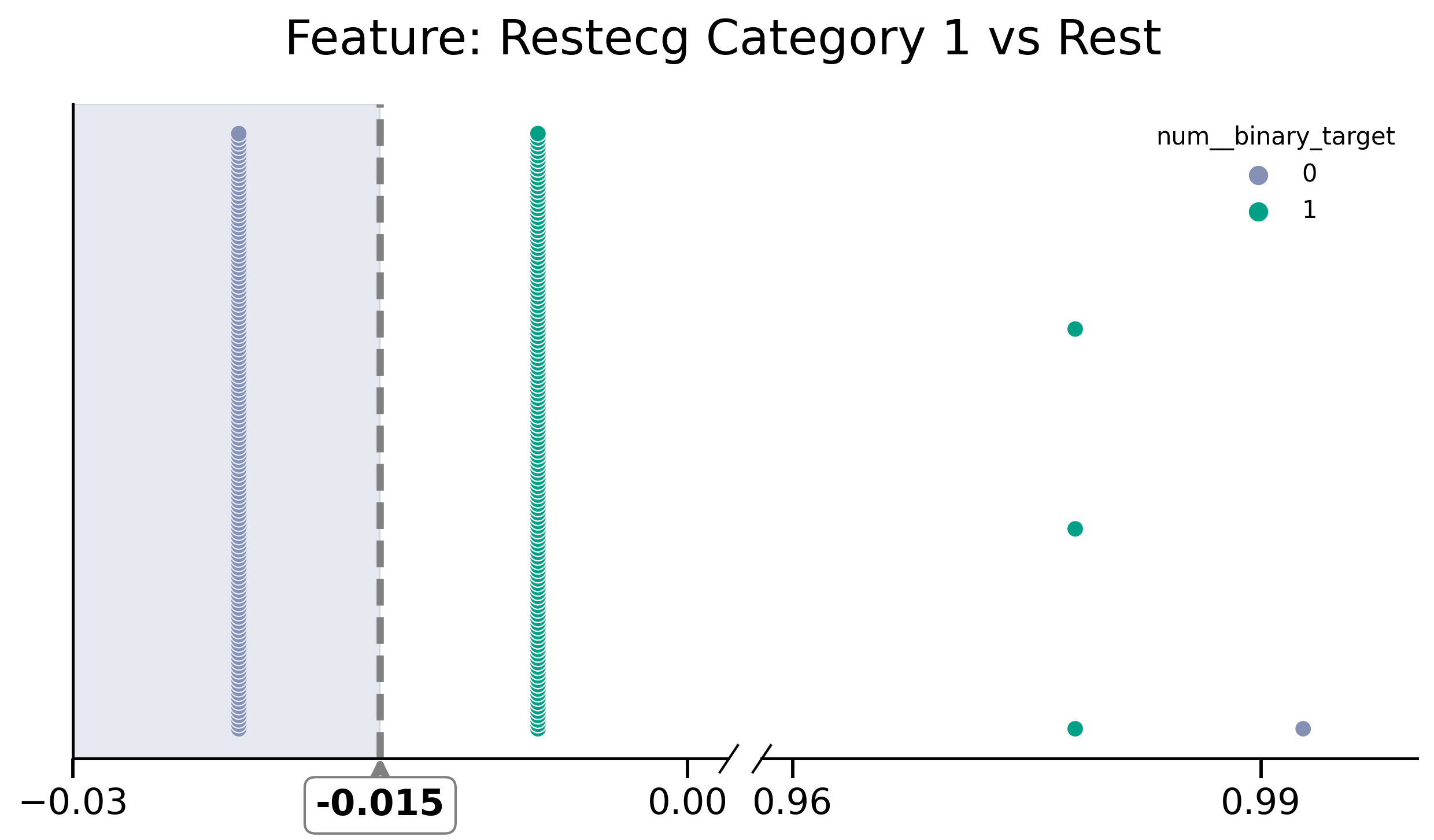
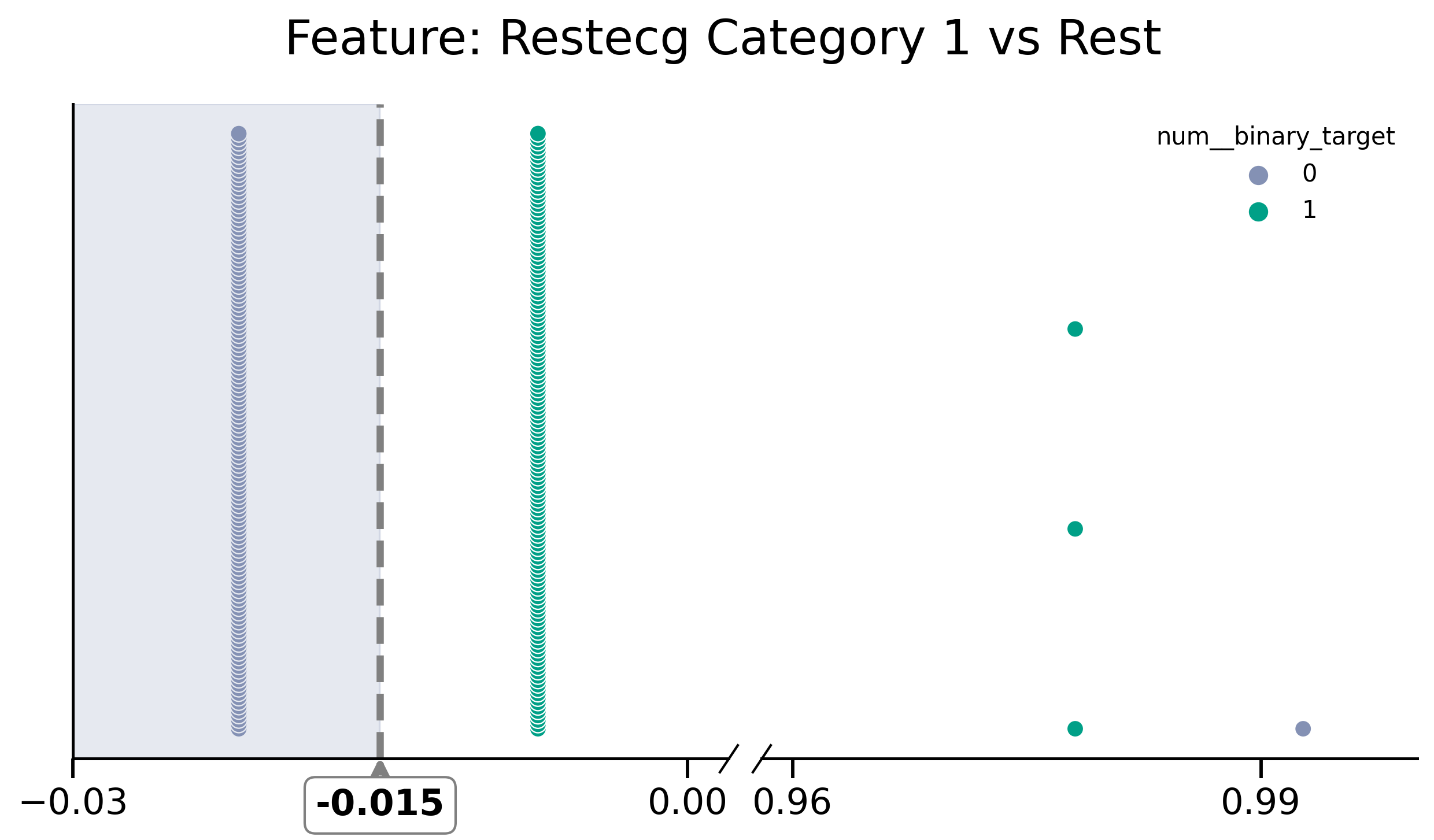
df_removed[y] = df[y]
fig, (ax1, ax2) = plot_discont(
"restecg_1__categorical",
y,
df_removed,
(-0.03, 0.002),
(0.958, 1),
colors=[green, purple],
title="Feature: Restecg Category 1 vs Rest",
)
print("threshold=", model_rm.named_steps.decisiontreeclassifier.tree_.threshold)
ax1.axvline(x=-0.015, color="grey", linestyle="dashed", lw=3)
ax1.axvspan(-0.03, -0.015, color=purple, alpha=0.2)
ax1.xaxis.set_major_locator(ticker.MultipleLocator(0.03))
ax2.xaxis.set_major_locator(ticker.MultipleLocator(0.03))
# visuals as we will make it smaller later we increase size
ax1.tick_params(axis="both", which="major", labelsize=15)
ax2.tick_params(axis="both", which="major", labelsize=15)
fig.suptitle("Feature: Restecg Category 1 vs Rest", size=20)
y_ground = ax1.transAxes.inverted().transform([0, 0])[1]
y_text = ax1.transAxes.inverted().transform([0, -0.1])[1]
ax1.annotate(
"-0.015",
xy=(-0.015, 0.01),
xytext=(-0.015, -0.045),
xycoords=ax1.get_xaxis_transform(),
verticalalignment="top",
ha="center",
arrowprops=dict(arrowstyle="->", color="grey", linewidth=3),
bbox=dict(boxstyle="round", fc="w", color="grey"),
fontsize=15,
weight="semibold",
)
fig.savefig("./saved_figures/heart_restecg__cat_1.svg")
fig
threshold= [-0.01459854 -2. -0.14233577 -2. -2. ]
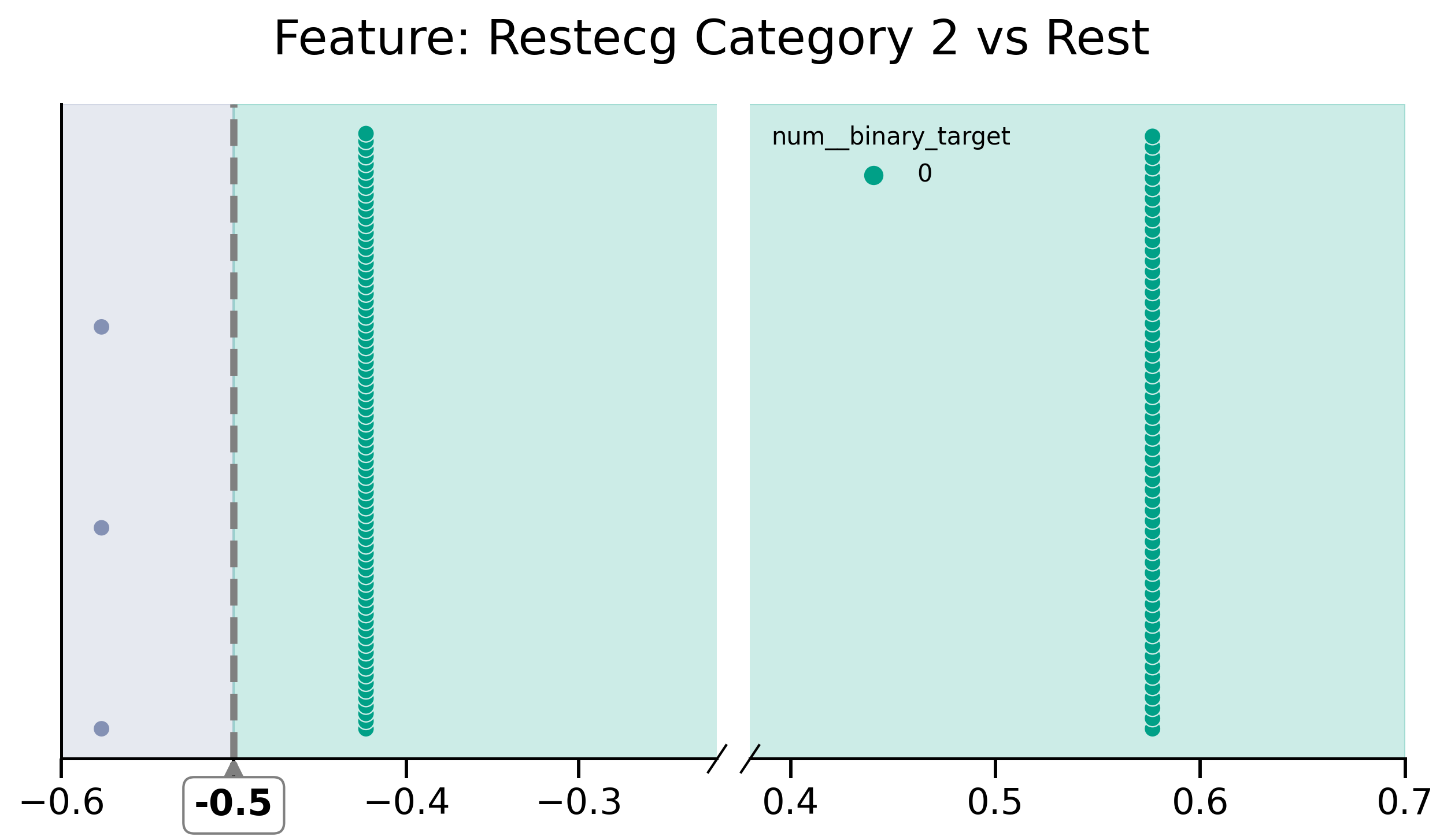
mask_selection = df_removed["restecg_1__categorical"] > -0.015
fig, (ax1, ax2) = plot_discont(
"restecg_2__categorical",
y,
df_removed[mask_selection],
(-0.6, -0.22),
(0.38, 0.7),
colors=[green, purple],
title="Feature: Restecg Category 2 vs Rest",
)
ax1.axvline(x=-0.5, color="grey", linestyle="--", lw=3)
ax1.axvspan(-0.6, -0.5, color=purple, alpha=0.2)
ax1.axvspan(-0.5, -0.1, color=green, alpha=0.2)
ax2.axvspan(0.35, 0.7, color=green, alpha=0.2)
ax1.xaxis.set_major_locator(ticker.MultipleLocator(0.1))
ax2.xaxis.set_major_locator(ticker.MultipleLocator(0.1))
# visuals as we will make it smaller later we increase size
ax1.tick_params(axis="both", which="major", labelsize=15)
ax2.tick_params(axis="both", which="major", labelsize=15)
fig.suptitle("Feature: Restecg Category 2 vs Rest", size=20)
ax1.annotate(
"-0.5",
xy=(-0.5, 0.01),
xytext=(-0.5, -0.045),
xycoords=ax1.get_xaxis_transform(),
verticalalignment="top",
ha="center",
arrowprops=dict(arrowstyle="->", color="grey", linewidth=3),
bbox=dict(boxstyle="round", fc="w", color="grey"),
fontsize=15,
weight="semibold",
)
fig.savefig("./saved_figures/heart_restecg__cat_2.svg")
Simulations: Impact of Small Changes in Features Leading to Leakage.¶
We only flip one feature value per fold to 1 or 0. Both taget and feature are binary. These simulations use only KFold for instead of RepeatedKFold
target = np.array([[0, 1] * 500]).flatten()
feat_1 = np.array([[0] * 2 + [1] * 2] * 250).flatten()
feat_2 = feat_1.copy()
for i, ii in zip(
[
97,
98,
197,
198,
297,
298,
397,
398,
497,
498,
597,
598,
697,
698,
797,
798,
897,
898,
997,
998,
],
[1, 0] * 10,
):
feat_2[i] = ii
df = pd.DataFrame(dict(feat_1=feat_1, feat_2=feat_2, conf=target, target=target))
np.random.seed(9464508)
scores_1 = run_cross_validation(
X=["feat_1"],
y="target",
data=df,
model=DecisionTreeClassifier(),
preprocess_X=["zscore"],
cv=KFold(10),
scoring="roc_auc",
)
scores_1 = scores_1.test_score
scores_2 = run_cross_validation(
X=["feat_2"],
y="target",
data=df,
model=DecisionTreeClassifier(),
preprocess_X=["zscore"],
cv=KFold(10),
scoring="roc_auc",
)
scores_2 = scores_2.test_score
print("No Removal:")
print(
"Scores Feat 1 (balanced): M = " f"{scores_1.mean():.4f}, sd = {scores_1.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"mean_carefully_constructed_balanced.txt",
scores_1.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"std_carefully_constructed_balanced.txt",
scores_1.std(),
)
print(
"Scores Feat 2 (conditional feature imbalance to TaCo in each fold): M = "
f"{scores_2.mean():.4f}, sd = {scores_2.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"mean_carefully_constructed.txt",
scores_2.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"raw",
"std_carefully_constructed.txt",
scores_2.std(),
)
No Removal:
Scores Feat 1 (balanced): M = 0.5000, sd = 0.0000
Scores Feat 2 (conditional feature imbalance to TaCo in each fold): M = 0.5200, sd = 0.0000
Now, we do the same but with TaCo removal:
results_1 = run_cross_validation(
X=["feat_1"],
confounds=["conf"],
y="target",
data=df,
model=DecisionTreeClassifier(),
preprocess_X=["zscore", "remove_confound"],
preprocess_confounds=["zscore"],
cv=KFold(10),
scoring="roc_auc",
)
score_1 = results_1.test_score
results_2 = run_cross_validation(
X=["feat_2"],
confounds=["conf"],
y="target",
data=df,
model=DecisionTreeClassifier(),
preprocess_X=["zscore", "remove_confound"],
preprocess_confounds=["zscore"],
cv=KFold(10),
scoring="roc_auc",
)
scores_2 = results_2.test_score
print(
"Scores Feat 1 (balanced): M = "
f"Mean = {scores_1.mean():.4f}, sd = {scores_1.std():.4f} \n"
"Scores Feat 2 (conditional feature imbalance to TaCo in each folds): "
f"Mean = {scores_2.mean():.4f}, sd = {scores_2.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_carefully_constructed_balanced.txt",
scores_1.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_carefully_constructed_balanced.txt",
scores_1.std(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"mean_carefully_constructed.txt",
scores_2.mean(),
)
save_paper_val(
base_save_paper,
"reasons_low_perc",
"removed",
"std_carefully_constructed.txt",
scores_2.std(),
)
Scores Feat 1 (balanced): M = Mean = 0.5000, sd = 0.0000
Scores Feat 2 (conditional feature imbalance to TaCo in each folds): Mean = 1.0000, sd = 0.0000