2a Reason: Continuous Features
Contents
2a Reason: Continuous Features¶
Imports
from pathlib import Path
from functools import partial
import numpy as np
import pandas as pd
from sklearn.model_selection import RepeatedStratifiedKFold, RepeatedKFold
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score
from sklearn.preprocessing import KBinsDiscretizer
import joblib
from scipy.stats import chi2
from julearn import run_cross_validation
import matplotlib.pyplot as plt
import matplotlib as mpl
from sciplotlib import style
import seaborn as sns
from leakconfound.plotting import mm_to_inch
from leakconfound.analyses.utils import save_paper_val
from leakconfound.plotting.settings import red, blue, green, purple
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
warnings.filterwarnings("ignore", category=FutureWarning)
Prepare Paths¶
base_dir = "../../"
base_save_paper = "./paper_val/"
increase_features_save_paper = "./paper_val/increase_features/"
Path(increase_features_save_paper).mkdir(parents=True, exist_ok=True)
Prepare Plotting¶
mpl.style.use(style.get_style("nature-reviews"))
mpl.rc("xtick", labelsize=11)
mpl.rc("ytick", labelsize=11)
mpl.rc("axes", labelsize=12, titlesize=12)
mpl.rc("figure", dpi=300)
mpl.rc("figure.subplot", wspace=mm_to_inch(4), hspace=mm_to_inch(10))
mpl.rc("lines", linewidth=1)
np.random.seed(891236740)
Functions to Sample from Distributions¶
def normal_with_bump(
loc_main, scale_main, loc_minor, scale_minor, size_main, size_minor
):
"""Function to fuse two normal distributions.
E.g. to create major normally distribution feature
then add another minor distribution around another value.
"""
arr_main = np.random.normal(loc_main, scale_main, size=size_main)
arr_minor = np.random.normal(loc_minor, scale_minor, size_minor)
arr_both = np.concatenate([arr_main, arr_minor])
np.random.shuffle(arr_both)
return arr_both
skewed_sampler = partial(chi2.rvs, df=3)
Features with Opposing Small Extreme Distributions¶
Prepare Data
target = np.array([[0, 1] * 500]).flatten()
feat_1 = np.zeros(len(target))
feat_1[target == 0] = normal_with_bump(0, 1, -4, 0.5, 450, 50)
feat_1[target == 1] = normal_with_bump(0, 1, 4, 0.5, 450, 50)
df = pd.DataFrame(dict(feat_1=feat_1, conf=target, target=target))
Run Analysis
res_no_rem, est_no_rem = run_cross_validation(
X=["feat_1"],
y="target",
data=df,
model=DecisionTreeClassifier(),
preprocess_X=["zscore"],
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
return_estimator="final",
)
scores_no_rem = res_no_rem.groupby("repeat").mean().test_score
res_rem, est_rem = run_cross_validation(
X=["feat_1"],
y="target",
data=df,
confounds=["conf"],
model=DecisionTreeClassifier(),
preprocess_X=["zscore", "remove_confound"],
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
return_estimator="final",
)
CV Scores
scores_rem = res_rem.groupby("repeat").mean().test_score
print(
f"Scores no removal: M = "
f"{scores_no_rem.mean():.4f}, sd = {scores_no_rem.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"raw",
"mean_opposing_extreme.txt",
round(scores_no_rem.mean(), 2),
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"raw",
"std_opposing_extreme.txt",
round(scores_no_rem.std(), 2),
)
print(
f"Scores with removal: M = " f"{scores_rem.mean():.4f}, sd = {scores_rem.std():.4f}"
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"removed",
"mean_opposing_extreme.txt",
round(scores_rem.mean(), 2),
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"removed",
"std_opposing_extreme.txt",
round(scores_rem.std(), 2),
)
Scores no removal: M = 0.5106, sd = 0.0107
Scores with removal: M = 0.5834, sd = 0.0130
Score on Additional Test Sample & Plotting¶
target = np.array([[0, 1] * 500]).flatten()
feat_1 = np.zeros(len(target))
feat_1[target == 0] = normal_with_bump(0, 1, -4, 0.5, 450, 50)
feat_1[target == 1] = normal_with_bump(0, 1, 4, 0.5, 450, 50)
df_test = pd.DataFrame(dict(feat_1=feat_1, conf=target, target=target))
pred_rm = est_rem.predict(df_test[["feat_1", "conf"]])
pred_no_rm = est_no_rem.predict(df_test[["feat_1"]])
score_rm = roc_auc_score(df_test["target"], pred_rm)
score_no_rm = roc_auc_score(df_test["target"], pred_no_rm)
print("New test set performance (same sampling procedure): ")
print(f"{score_rm = } \n {score_no_rm = }")
New test set performance (same sampling procedure):
score_rm = 0.585
score_no_rm = 0.534
X_rm, _ = est_rem.preprocess(df_test[["feat_1", "conf"]], df_test.target)
X_no_rm, _ = est_no_rem.preprocess(df_test[["feat_1"]], df_test.target)
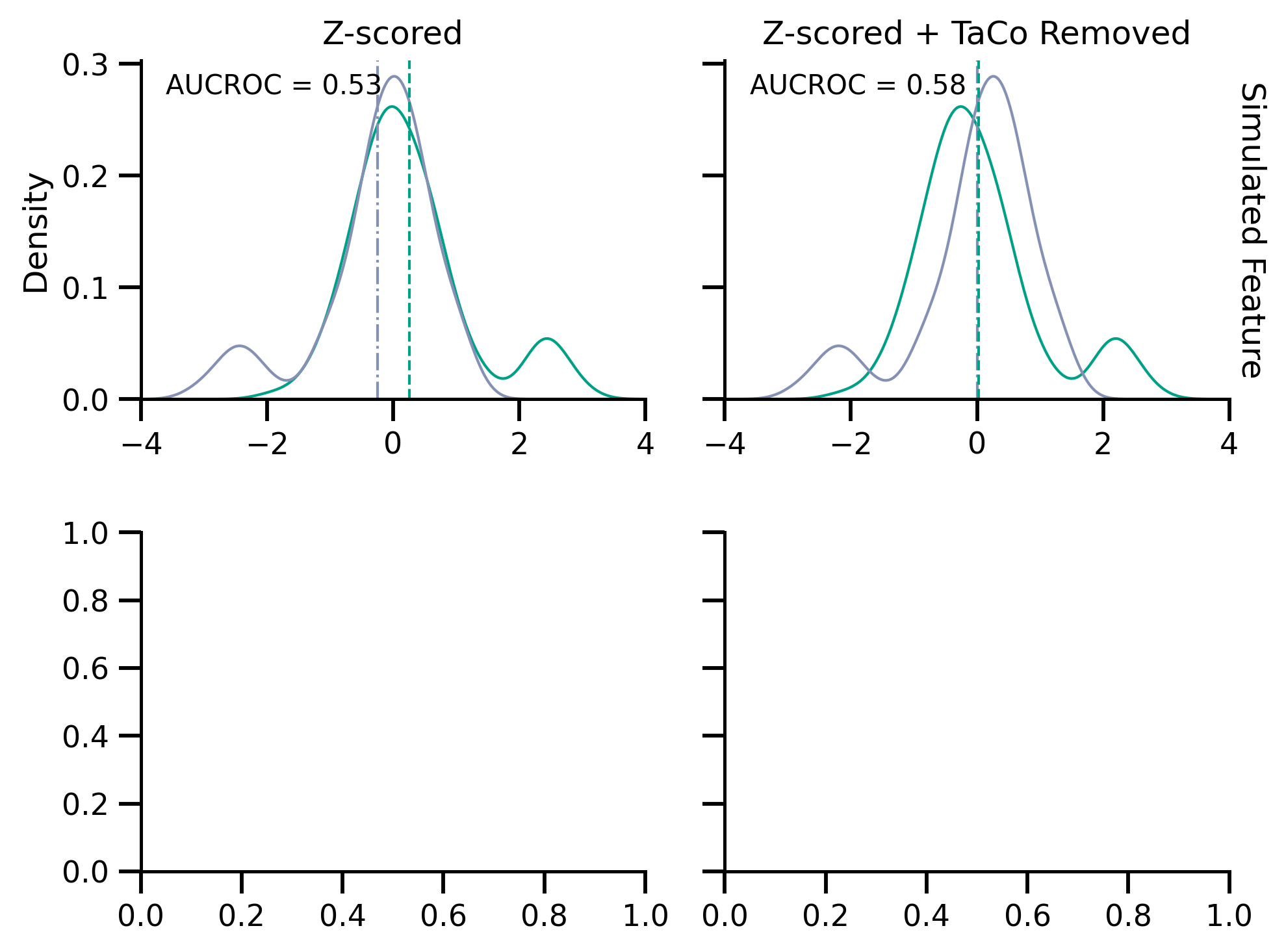
fig, axes = plt.subplots(2, 2, figsize=[mm_to_inch(183), mm_to_inch(140)], sharey="row")
g0 = sns.kdeplot(
x=X_no_rm.feat_1,
hue=df_test.target,
ax=axes[0, 0],
palette=sns.color_palette([purple, green]),
)
g0.set_xlim(-4, 4)
g = sns.kdeplot(
x=X_rm.feat_1,
hue=df_test.target,
ax=axes[0, 1],
palette=sns.color_palette([purple, green]),
)
g.set_xlim(-4, 4)
axes[0, 0].axvline(
X_no_rm.feat_1[df_test.target == 0].values.mean(), color=purple, ls="-."
)
axes[0, 0].axvline(
X_no_rm.feat_1[df_test.target == 1].values.mean(), color=green, ls="--"
)
axes[0, 1].axvline(
X_rm.feat_1[df_test.target == 0].values.mean(), color=purple, ls="-."
)
axes[0, 1].axvline(X_rm.feat_1[df_test.target == 1].values.mean(), color=green, ls="--")
ax_twin = axes[0, 1].twinx()
ax_twin.set_ylabel(
"Simulated Feature",
rotation=270,
labelpad=8,
horizontalalignment="center",
verticalalignment="center",
)
ax_twin.set_yticks([])
ax_twin.set_yticklabels("")
for ax in axes.flatten():
ax.set_xlabel("")
axes[0, 0].set_title("Z-scored")
axes[0, 1].set_title("Z-scored + TaCo Removed")
axes[0, 0].get_legend().set_visible(False)
axes[0, 1].get_legend().set_visible(False)
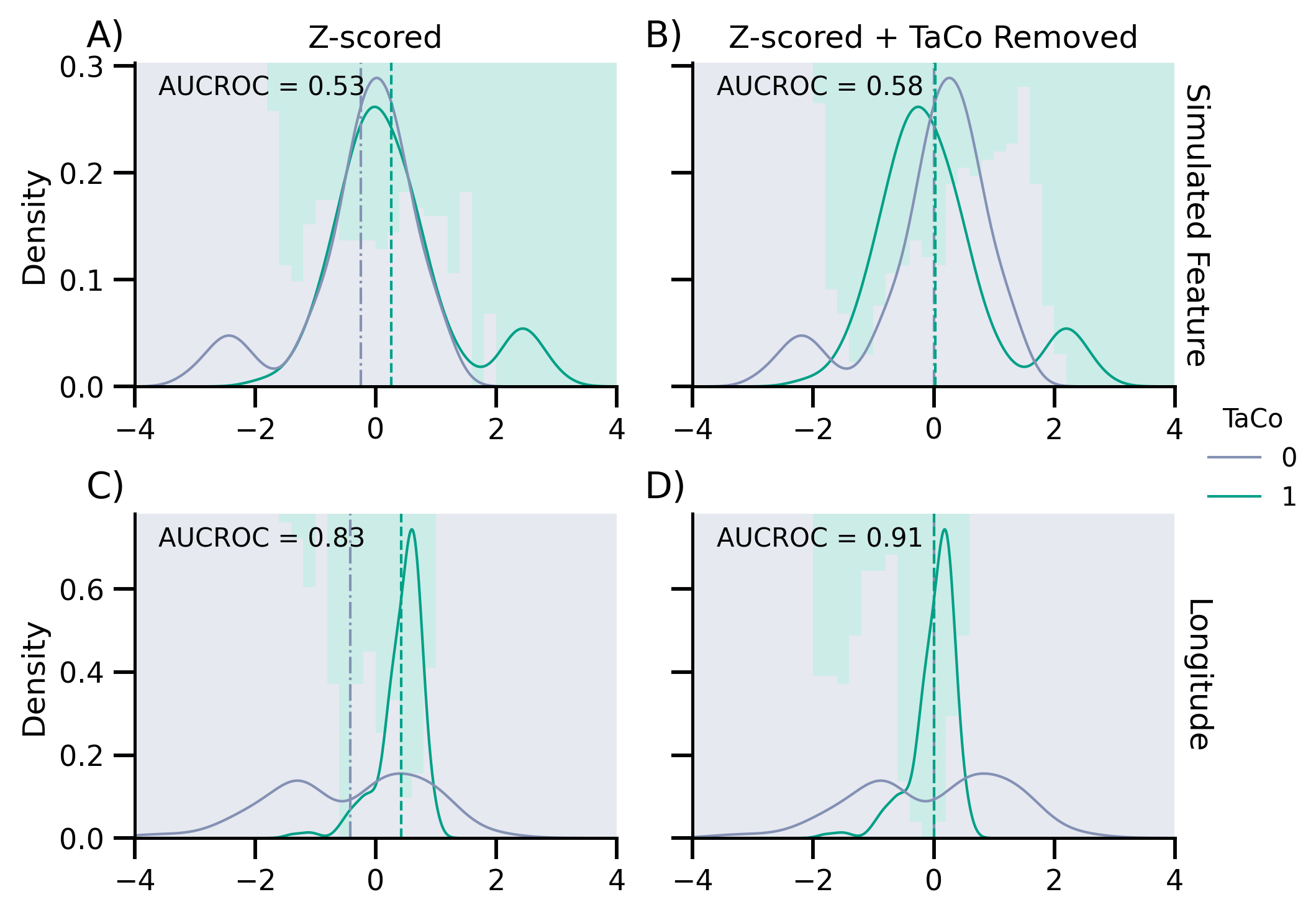
axes[0, 0].text(0.05, 0.9, f"AUCROC = {score_no_rm:.2}", transform=axes[0, 0].transAxes)
axes[0, 1].text(0.05, 0.9, f"AUCROC = {score_rm:.2}", transform=axes[0, 1].transAxes)
Text(0.05, 0.9, 'AUCROC = 0.58')
def plot_bin(row, ax):
x_min = row.name.left
x_max = row.name.right
changing_point = 1 - row.values[0]
ax.axvspan(
x_min,
x_max,
0,
changing_point,
facecolor=purple,
alpha=0.2,
linewidth=None,
ls="",
)
ax.axvspan(
x_min,
x_max,
changing_point,
1,
facecolor=green,
alpha=0.2,
linewidth=None,
ls="",
)
bin_number = 40
x_background_rm = np.arange(-4, 4, 0.005)
x_background_no_rm = np.arange(-4, 4, 0.005)
pred_all_val_no_rm = est_no_rem.named_steps.decisiontreeclassifier.predict(
pd.DataFrame(dict(feat_1=x_background_no_rm))
)
(
pd.DataFrame(dict(pred=pred_all_val_no_rm, x=x_background_no_rm))
.assign(bins=lambda df: pd.qcut(df.x, bin_number))
.pipe(lambda df: pd.DataFrame(df.groupby("bins").pred.mean()))
.apply(plot_bin, axis=1, ax=axes[0, 0])
)
pred_all_val_rm = est_rem.named_steps.decisiontreeclassifier.predict(
pd.DataFrame(dict(feat_1=x_background_rm))
)
(
pd.DataFrame(dict(pred=pred_all_val_rm, x=x_background_rm))
.assign(bins=lambda df: pd.qcut(df.x, bin_number))
.pipe(lambda df: pd.DataFrame(df.groupby("bins").pred.mean()))
.apply(plot_bin, axis=1, ax=axes[0, 1])
)
bins
(-4.001, -3.8] None
(-3.8, -3.6] None
(-3.6, -3.4] None
(-3.4, -3.201] None
(-3.201, -3.001] None
(-3.001, -2.801] None
(-2.801, -2.601] None
(-2.601, -2.401] None
(-2.401, -2.201] None
(-2.201, -2.001] None
(-2.001, -1.801] None
(-1.801, -1.602] None
(-1.602, -1.402] None
(-1.402, -1.202] None
(-1.202, -1.002] None
(-1.002, -0.802] None
(-0.802, -0.602] None
(-0.602, -0.402] None
(-0.402, -0.202] None
(-0.202, -0.0025] None
(-0.0025, 0.197] None
(0.197, 0.397] None
(0.397, 0.597] None
(0.597, 0.797] None
(0.797, 0.997] None
(0.997, 1.197] None
(1.197, 1.397] None
(1.397, 1.596] None
(1.596, 1.796] None
(1.796, 1.996] None
(1.996, 2.196] None
(2.196, 2.396] None
(2.396, 2.596] None
(2.596, 2.796] None
(2.796, 2.996] None
(2.996, 3.195] None
(3.195, 3.395] None
(3.395, 3.595] None
(3.595, 3.795] None
(3.795, 3.995] None
dtype: object
Score on Additional Test Sample Without Extreme Distributions¶
Testing performance on addition test set only using a normally distributed features. This shows whether increase in performance is due to leakage or only better predictions of extreme values.
target = np.array([[0, 1] * 500]).flatten()
feat_1 = np.zeros(len(target))
feat_1[target == 0] = np.random.normal(size=500)
feat_1[target == 1] = np.random.normal(size=500)
df_test = pd.DataFrame(dict(feat_1=feat_1, conf=target, target=target))
pred_rm = est_rem.predict(df_test[["feat_1", "conf"]])
pred_no_rm = est_no_rem.predict(df_test[["feat_1"]])
score_rm = roc_auc_score(df_test["target"], pred_rm)
score_no_rm = roc_auc_score(df_test["target"], pred_no_rm)
print("New test set performance (sampling only normal dist): ")
print(f"{score_rm = } \n {score_no_rm = }")
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"raw",
"mean_opposing_extreme_extra_test.txt",
score_no_rm.mean(),
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"removed",
"mean_opposing_extreme_extra_test.txt",
score_rm.mean(),
)
New test set performance (sampling only normal dist):
score_rm = 0.5650000000000001
score_no_rm = 0.484
Real World Example¶
Now we can look at an example with real data: Therefore, we will look at the real_estate dataset.
# read in the models and the input data
case_rm = "real_estate___decisiontree___True___716845___house_price_of_unit_area"
case_none = "real_estate___decisiontree___False___716845___house_price_of_unit_area"
decisiontree_rm = joblib.load(
f"{base_dir}/results/basic_TaCo/uci_datasets/models/{case_rm}.joblib"
)["decisiontreeregressor"]
decisiontree_none = joblib.load(
f"{base_dir}/results/basic_TaCo/uci_datasets/models/{case_none}.joblib"
)["decisiontreeregressor"]
df = pd.read_csv(f"{base_dir}data/uci_datasets/real_estate.csv")
X = [
col
for col in df.columns.to_list()
if col.endswith("categorical") or col.endswith("continuous")
]
confounds = ["house_price_of_unit_area__:type:__confound"]
y = "house_price_of_unit_area__regression_target"
df[confounds[0]] = df[y] # make it TaCo
First, let us have a look at which features were important in the original prediction task with and without TaCo removal
print("importance with removal")
print(decisiontree_rm.feature_importances_)
print("most important variables are 1, 4:", df.iloc[:, [1, 4]].columns)
print()
print("importance without removal")
print(decisiontree_none.feature_importances_)
importance with removal
[0.1055301 0.34810974 0.07908374 0.07494882 0.3923276 ]
most important variables are 1, 4: Index(['distance_to_the_nearest_MRT_station__continuous', 'longitude__continuous'], dtype='object')
importance without removal
[0.19743099 0.61044498 0.02933386 0.09701904 0.06577114]
Now, we retrain and score a decision tree using only these features on a binarized TaCo for interpretation purposes.
discretizer = KBinsDiscretizer(n_bins=2, strategy="quantile", encode="ordinal").fit(
df[[y]]
)
y_ = "House Price Quantiles"
df[y_] = discretizer.transform(df[[y]]).reshape(-1)
score_no_rm, model_bin_no_rm = run_cross_validation(
X=["longitude__continuous"],
preprocess_X=["zscore"],
y=y_,
data=df,
model=DecisionTreeClassifier(),
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
return_estimator="final",
)
pred_no_rm = model_bin_no_rm.predict(df[["longitude__continuous"]])
score_rm, model_bin_rm = run_cross_validation(
X=["longitude__continuous"],
preprocess_X=["zscore", "remove_confound"],
confounds=[y_],
y=y_,
data=df,
model=DecisionTreeClassifier(),
cv=RepeatedStratifiedKFold(),
scoring="roc_auc",
return_estimator="final",
)
m_rm = score_rm.groupby("repeat").mean().test_score
m_no_rm = score_no_rm.groupby("repeat").mean().test_score
print("Scores DT pred binarized House Prize given longitude ")
print(f"Scores no removal: M = " f"{m_no_rm.mean():.4f}, sd = {m_no_rm.std():.4f}")
print(f"Scores with removal: M = " f"{m_rm.mean():.4f}, sd = {m_rm.std():.4f}")
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"raw",
"mean_dt_binarized_house.txt",
round(m_no_rm.mean(), 2),
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"raw",
"std_dt_binarized_house.txt",
round(m_no_rm.std(), 2),
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"removed",
"mean_dt_binarized_house.txt",
round(m_rm.mean(), 2),
)
save_paper_val(
base_save_paper,
"reasons_continuous_feat",
"removed",
"std_dt_binarized_house.txt",
round(m_rm.std(), 2),
)
Scores DT pred binarized House Prize given longitude
Scores no removal: M = 0.8324, sd = 0.0100
Scores with removal: M = 0.9080, sd = 0.0135
pred_rm = model_bin_rm.predict(df[["longitude__continuous", y_]])
X_rm, _ = model_bin_rm.preprocess(df[["longitude__continuous", y_]], df[y_])
X_no_rm, _ = model_bin_no_rm.preprocess(df[["longitude__continuous"]], df[y_])
g0 = sns.kdeplot(
x=X_no_rm["longitude__continuous"],
hue=df[y_],
ax=axes[1, 0],
palette=sns.color_palette([purple, green]),
)
axes[1, 0].axvline(X_no_rm[df[y_] == 0].values.mean(), color=purple, ls="-.")
axes[1, 0].axvline(X_no_rm[df[y_] == 1].values.mean(), color=green, ls="--")
axes[1, 1].axvline(X_rm[df[y_] == 0].values.mean(), color=purple, ls="-.")
axes[1, 1].axvline(X_rm[df[y_] == 1].values.mean(), color=green, ls="--")
g0.set_xlim(-4, 4)
g = sns.kdeplot(
x=X_rm["longitude__continuous"],
hue=df[y_],
ax=axes[1, 1],
palette=sns.color_palette([purple, green]),
)
g.set_xlim(-4, 4)
ax_twin = axes[1, 1].twinx()
ax_twin.set_ylabel(
"Longitude",
rotation=270,
labelpad=8,
horizontalalignment="center",
verticalalignment="center",
)
ax_twin.set_yticks([])
ax_twin.set_yticklabels("")
axes[1, 0].set_title("")
axes[1, 1].set_title("")
axes[1, 0].get_legend().set_visible(False)
axes[1, 1].get_legend().set_visible(False)
axes[1, 0].text(
0.05,
0.9,
f"AUCROC = {score_no_rm.test_score.mean() :.2}",
transform=axes[1, 0].transAxes,
)
axes[1, 1].text(
0.05,
0.9,
f"AUCROC = {score_rm.test_score.mean():.2}",
transform=axes[1, 1].transAxes,
)
for ax in axes[1]:
ax.set_xlabel("")
bin_number = 40
x_background = np.arange(-4, 4, 0.005)
pred_all_val_no_rm = model_bin_no_rm.named_steps.decisiontreeclassifier.predict(
pd.DataFrame(dict(longitude__continuous=x_background))
)
(
pd.DataFrame(dict(pred=pred_all_val_no_rm, x=x_background))
.assign(bins=lambda df: pd.qcut(df.x, bin_number))
.pipe(lambda df: pd.DataFrame(df.groupby("bins").pred.mean()))
.apply(plot_bin, axis=1, ax=axes[1, 0])
)
pred_all_val_rm = model_bin_rm.named_steps.decisiontreeclassifier.predict(
pd.DataFrame(dict(longitude__continuous=x_background))
)
(
pd.DataFrame(dict(pred=pred_all_val_rm, x=x_background))
.assign(bins=lambda df: pd.qcut(df.x, bin_number))
.pipe(lambda df: pd.DataFrame(df.groupby("bins").pred.mean()))
.apply(plot_bin, axis=1, ax=axes[1, 1])
)
bins
(-4.001, -3.8] None
(-3.8, -3.6] None
(-3.6, -3.4] None
(-3.4, -3.201] None
(-3.201, -3.001] None
(-3.001, -2.801] None
(-2.801, -2.601] None
(-2.601, -2.401] None
(-2.401, -2.201] None
(-2.201, -2.001] None
(-2.001, -1.801] None
(-1.801, -1.602] None
(-1.602, -1.402] None
(-1.402, -1.202] None
(-1.202, -1.002] None
(-1.002, -0.802] None
(-0.802, -0.602] None
(-0.602, -0.402] None
(-0.402, -0.202] None
(-0.202, -0.0025] None
(-0.0025, 0.197] None
(0.197, 0.397] None
(0.397, 0.597] None
(0.597, 0.797] None
(0.797, 0.997] None
(0.997, 1.197] None
(1.197, 1.397] None
(1.397, 1.596] None
(1.596, 1.796] None
(1.796, 1.996] None
(1.996, 2.196] None
(2.196, 2.396] None
(2.396, 2.596] None
(2.596, 2.796] None
(2.796, 2.996] None
(2.996, 3.195] None
(3.195, 3.395] None
(3.395, 3.595] None
(3.595, 3.795] None
(3.795, 3.995] None
dtype: object
handles = [
mpl.lines.Line2D([], [], color=purple, label="0"),
mpl.lines.Line2D([], [], color=green, label="1"),
]
fig.legend(handles=handles, title="TaCo", loc=[0.91, 0.43])
for ax, txt in zip(axes.flatten(), ["A)", "B)", "C)", "D)"]):
ax.text(-0.1, 1.05, txt, transform=ax.transAxes, size=14)
fig
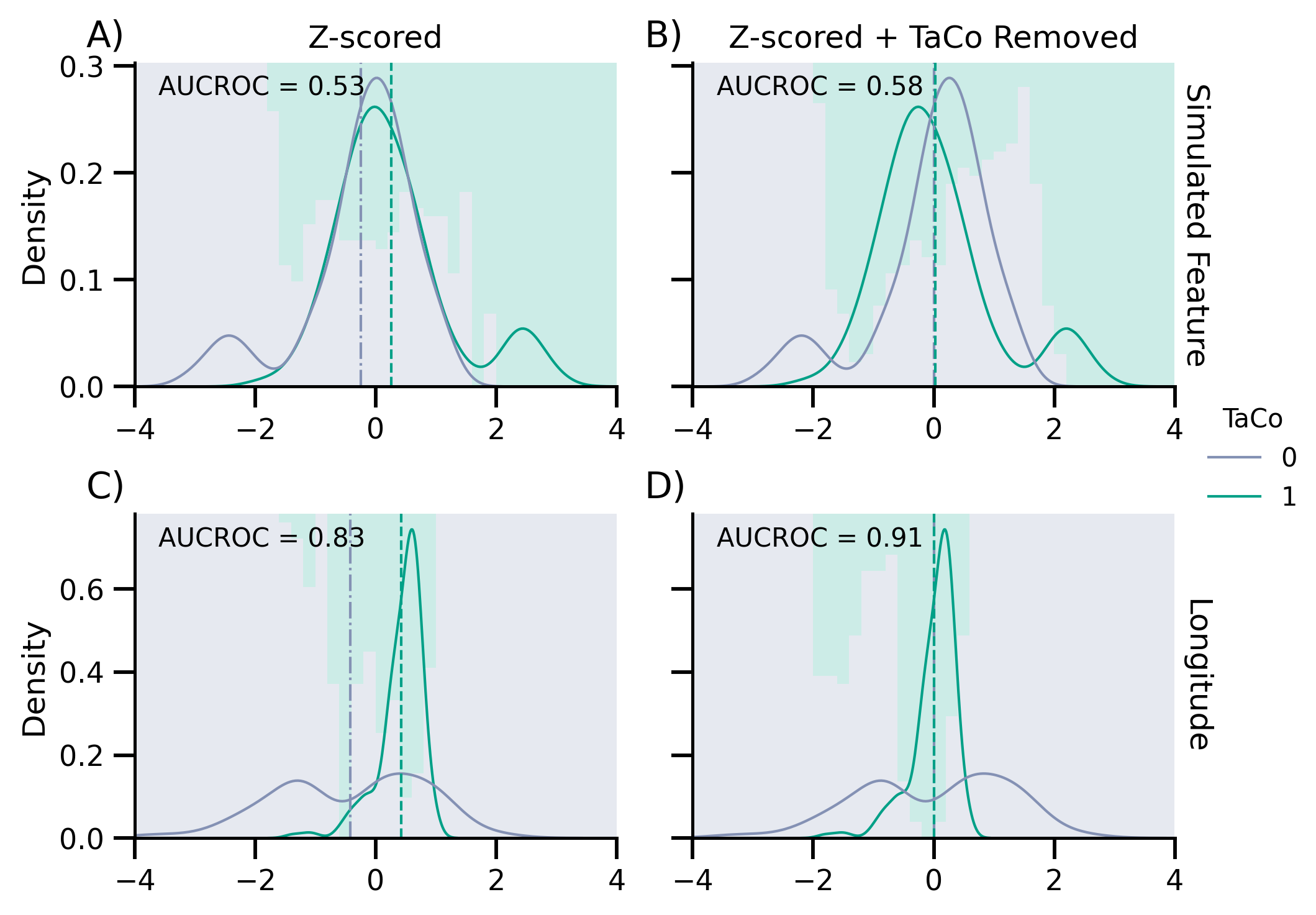
fig.savefig("./saved_figures/03_binary_complete.svg")
fig.savefig("./saved_figures/03_binary_complete.png")
joblib.dump([fig, axes], "./saved_figures/03_binary_complete.figure")
fig
Analysing The Effect of Random Features¶
using either normally distributed or following chi2 features.
target = np.random.normal(size=500)
df = pd.DataFrame(dict(conf=target, target=target))
for i in range(100):
df[f"feat_{i}"] = skewed_sampler(size=500)
/tmp/ipykernel_12609/3003596449.py:5: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
df[f"feat_{i}"] = skewed_sampler(size=500)
First using normally distributed random features:
all_scores_no_rem = []
all_yerr_no_rem = []
all_scores_rem = []
all_yerr_rem = []
feat_nbr = [1, *list(range(10, 101, 10))]
for i in feat_nbr:
scores_no_rem = run_cross_validation(
X=[f"feat_{ii}" for ii in range(i)],
y="target",
data=df,
model="rf",
preprocess_confounds="zscore",
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
)
scores_no_rem = scores_no_rem.groupby("repeat")["test_score"].mean()
all_scores_no_rem.append(scores_no_rem.mean())
all_yerr_no_rem.append(scores_no_rem.std())
scores_rem = run_cross_validation(
X=[f"feat_{ii}" for ii in range(i)],
y="target",
data=df,
confounds=["conf"],
model="rf",
preprocess_X=["zscore", "remove_confound"],
preprocess_confounds="zscore",
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
)
scores_rem = scores_rem.groupby("repeat")["test_score"].mean()
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"raw",
f"mean_iter_{i}.txt",
scores_no_rem.mean(),
)
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"raw",
f"std_iter_{i}.txt",
scores_no_rem.std(),
)
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"removed",
f"mean_iter_{i}.txt",
scores_rem.mean(),
)
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"removed",
f"std_iter_{i}.txt",
scores_rem.std(),
)
all_scores_rem.append(scores_rem.mean())
all_yerr_rem.append(scores_rem.std())
df_rem = pd.DataFrame(
dict(scores=all_scores_rem, iteration=feat_nbr, yerr=all_yerr_rem)
)
df_rem["remove confounds"] = "confound removed"
df_no_rem = pd.DataFrame(
dict(scores=all_scores_no_rem, iteration=feat_nbr, yerr=all_yerr_no_rem)
)
df_no_rem["remove confounds"] = "no removal"
df_plot = pd.concat([df_rem, df_no_rem])
df_plot["confound"] = df_plot["remove confounds"].apply(
lambda x: ("removed" if x == "confound removed" else "not removed")
)
fig, ax = plt.subplots(figsize=[mm_to_inch(180), mm_to_inch(40)])
g = sns.lineplot(
x="iteration",
y="scores",
hue="confound",
hue_order=["not removed", "removed"],
data=df_plot.reset_index(),
palette=sns.color_palette([blue, red]),
ax=ax,
)
g.set(xlabel="Number Of Features", ylabel="R²", title="χ2 Distributed Features")
df_plot_removed = df_plot.query('confound=="removed"')
ax.errorbar(
df_plot_removed["iteration"],
df_plot_removed["scores"],
df_plot_removed["yerr"],
fmt="none",
color=red,
)
df_plot_not_removed = df_plot.query('confound=="not removed"')
ax.errorbar(
df_plot_not_removed["iteration"],
df_plot_not_removed["scores"],
df_plot_not_removed["yerr"],
fmt="none",
color=blue,
)
ax.yaxis.set_major_locator(mpl.ticker.FixedLocator([-0.4, -0.2, 0, 0.2]))
ax.yaxis.set_major_formatter(mpl.ticker.FixedFormatter([-0.4, -0.2, 0, 0.2]))
ax.set_ylim(-0.4, 0.25)
fig.savefig("./saved_figures/increasing_feat_chi.png")
fig.savefig("./saved_figures/increasing_feat_chi.svg")
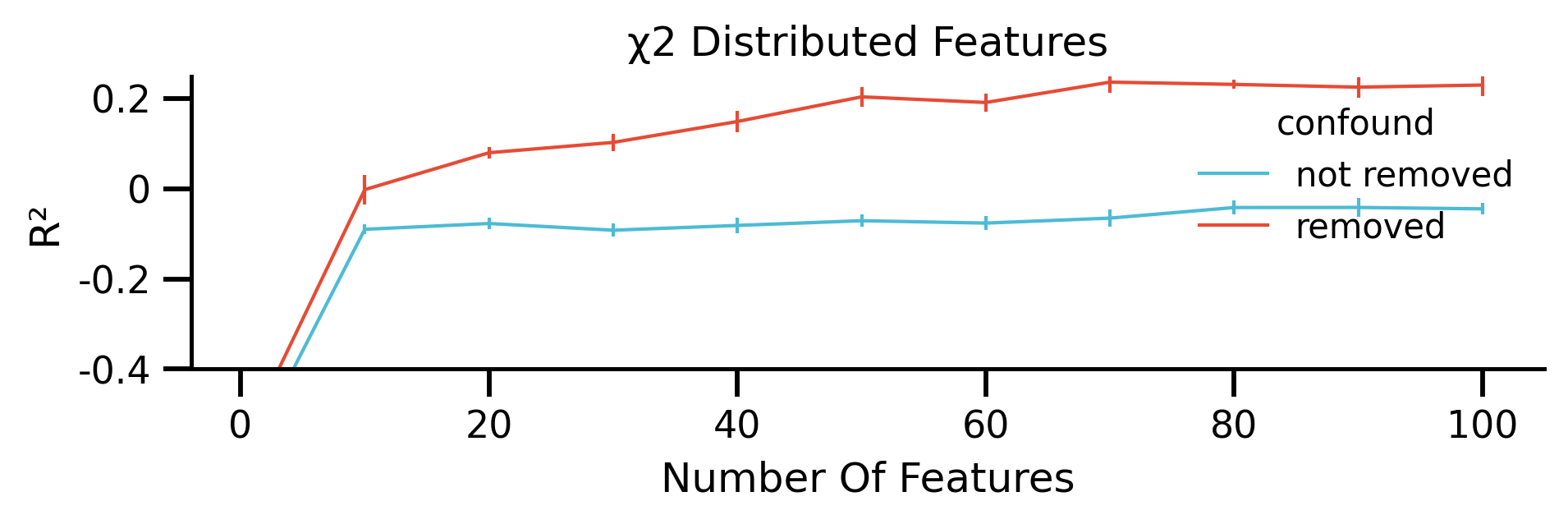
Same for not skewed features:
target = np.random.normal(size=500)
df = pd.DataFrame(dict(conf=target, target=target))
for i in range(100):
df[f"feat_{i}"] = np.random.normal(size=500)
/tmp/ipykernel_12609/2537863411.py:5: PerformanceWarning: DataFrame is highly fragmented. This is usually the result of calling `frame.insert` many times, which has poor performance. Consider joining all columns at once using pd.concat(axis=1) instead. To get a de-fragmented frame, use `newframe = frame.copy()`
df[f"feat_{i}"] = np.random.normal(size=500)
all_scores_no_rem = []
all_yerr_no_rem = []
all_scores_rem = []
all_yerr_rem = []
for i in feat_nbr:
scores_no_rem = run_cross_validation(
X=[f"feat_{ii}" for ii in range(i)],
y="target",
data=df,
model="rf",
preprocess_confounds="zscore",
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
)
scores_no_rem = scores_no_rem.groupby("repeat")["test_score"].mean()
all_scores_no_rem.append(scores_no_rem.mean())
all_yerr_no_rem.append(scores_no_rem.std())
scores_rem = run_cross_validation(
X=[f"feat_{ii}" for ii in range(i)],
y="target",
data=df,
confounds=["conf"],
model="rf",
preprocess_X=["zscore", "remove_confound"],
preprocess_confounds="zscore",
cv=RepeatedKFold(),
scoring="r2",
problem_type="regression",
)
scores_rem = scores_rem.groupby("repeat")["test_score"].mean()
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"raw",
"mean_increasing_features.txt",
scores_no_rem.mean(),
)
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"raw",
"std_increasing_features.txt",
scores_no_rem.std(),
)
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"removed",
"mean_increasing_features.txt",
scores_rem.mean(),
)
save_paper_val(
increase_features_save_paper,
"reasons_continuous_feat",
"removed",
"std_increasing_features.txt",
scores_rem.std(),
)
all_scores_rem.append(scores_rem.mean())
all_yerr_rem.append(scores_rem.std())
df_rem = pd.DataFrame(
dict(scores=all_scores_rem, iteration=feat_nbr, yerr=all_yerr_rem)
)
df_rem["remove confounds"] = "confound removed"
df_no_rem = pd.DataFrame(
dict(scores=all_scores_no_rem, iteration=feat_nbr, yerr=all_yerr_no_rem)
)
df_no_rem["remove confounds"] = "no removal"
df_plot = pd.concat([df_rem, df_no_rem])
df_plot["confound"] = df_plot["remove confounds"].apply(
lambda x: ("removed" if x == "confound removed" else "not removed")
)
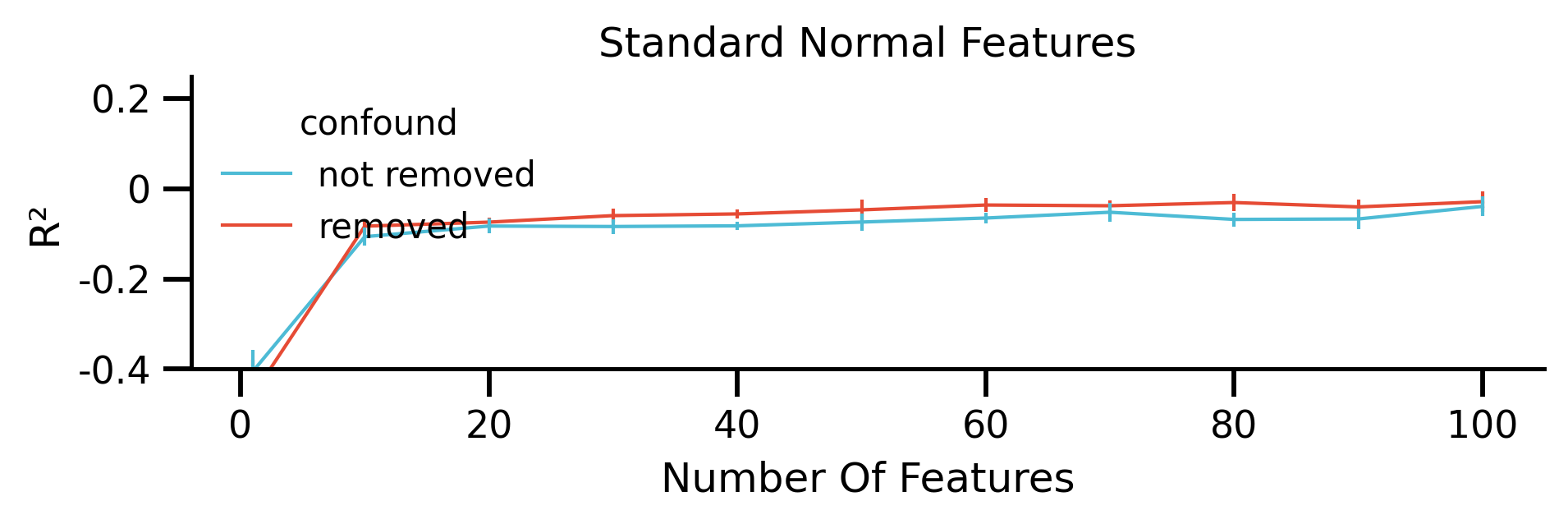
fig, ax = plt.subplots(figsize=[mm_to_inch(180), mm_to_inch(40)])
g = sns.lineplot(
x="iteration",
y="scores",
hue="confound",
hue_order=["not removed", "removed"],
data=df_plot.reset_index(),
palette=sns.color_palette([blue, red]),
ax=ax,
)
g.set(xlabel="Number Of Features", ylabel="R²", title="Standard Normal Features")
df_plot_removed = df_plot.query('confound=="removed"')
ax.errorbar(
df_plot_removed["iteration"],
df_plot_removed["scores"],
df_plot_removed["yerr"],
fmt="none",
color=red,
)
df_plot_not_removed = df_plot.query('confound=="not removed"')
ax.errorbar(
df_plot_not_removed["iteration"],
df_plot_not_removed["scores"],
df_plot_not_removed["yerr"],
fmt="none",
color=blue,
)
ax.yaxis.set_major_locator(mpl.ticker.FixedLocator([-0.4, -0.2, 0, 0.2]))
ax.yaxis.set_major_formatter(mpl.ticker.FixedFormatter([-0.4, -0.2, 0, 0.2]))
ax.set_ylim(-0.4, 0.25)
fig.savefig("./saved_figures/increasing_feat_normal.png")
fig.savefig("./saved_figures/increasing_feat_normal.svg")