Note
Click here to download the full example code
Regression Analysis¶
This example uses the ‘diabetes’ data from sklearn datasets and performs a regression analysis using a Ridge Regression model.
# Authors: Shammi More <s.more@fz-juelich.de>
# Federico Raimondo <f.raimondo@fz-juelich.de>
#
# License: AGPL
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_diabetes
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from julearn import run_cross_validation
from julearn.utils import configure_logging
Out:
/home/travis/virtualenv/python3.7.1/lib/python3.7/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
return f(*args, **kwds)
Set the logging level to info to see extra information
configure_logging(level='INFO')
Out:
2021-01-28 20:06:57,741 - julearn - INFO - ===== Lib Versions =====
2021-01-28 20:06:57,741 - julearn - INFO - numpy: 1.19.5
2021-01-28 20:06:57,741 - julearn - INFO - scipy: 1.6.0
2021-01-28 20:06:57,741 - julearn - INFO - sklearn: 0.24.1
2021-01-28 20:06:57,741 - julearn - INFO - pandas: 1.2.1
2021-01-28 20:06:57,741 - julearn - INFO - julearn: 0.2.5.dev19+g9c15c5f
2021-01-28 20:06:57,741 - julearn - INFO - ========================
load the diabetes data from sklearn as a pandas dataframe
features, target = load_diabetes(return_X_y=True, as_frame=True)
Dataset contains ten variables age, sex, body mass index, average blood pressure, and six blood serum measurements (s1-s6) diabetes patients and a quantitative measure of disease progression one year after baseline which is the target we are interested in predicting.
print('Features: \n', features.head())
print('Target: \n', target.describe())
Out:
Features:
age sex bmi ... s4 s5 s6
0 0.038076 0.050680 0.061696 ... -0.002592 0.019908 -0.017646
1 -0.001882 -0.044642 -0.051474 ... -0.039493 -0.068330 -0.092204
2 0.085299 0.050680 0.044451 ... -0.002592 0.002864 -0.025930
3 -0.089063 -0.044642 -0.011595 ... 0.034309 0.022692 -0.009362
4 0.005383 -0.044642 -0.036385 ... -0.002592 -0.031991 -0.046641
[5 rows x 10 columns]
Target:
count 442.000000
mean 152.133484
std 77.093005
min 25.000000
25% 87.000000
50% 140.500000
75% 211.500000
max 346.000000
Name: target, dtype: float64
Let’s combine features and target together in one dataframe and define X and y
calculate correlations between the features/variables and plot it as heat map
corr = data_diabetes.corr()
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.set(font_scale=1.2)
sns.heatmap(corr, xticklabels=corr.columns, yticklabels=corr.columns,
annot=True, fmt="0.1f")
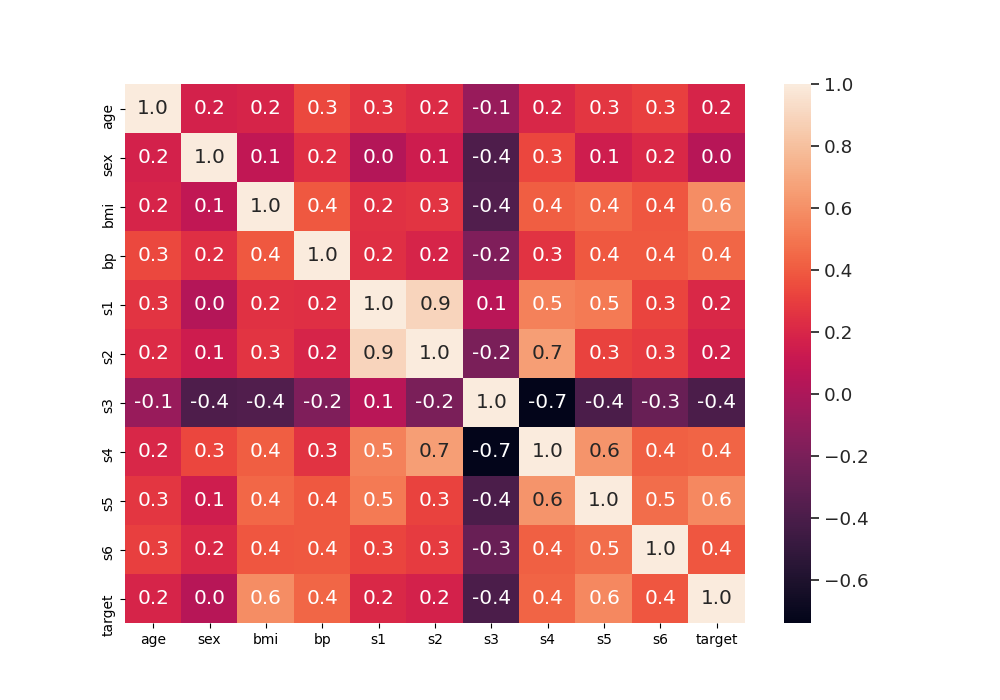
Out:
<AxesSubplot:>
Split the dataset into train and test
train_diabetes, test_diabetes = train_test_split(data_diabetes, test_size=0.3)
Train a ridge regression model on train dataset and use mean absolute error for scoring
Out:
2021-01-28 20:06:58,158 - julearn - INFO - Using default CV
2021-01-28 20:06:58,159 - julearn - INFO - ==== Input Data ====
2021-01-28 20:06:58,159 - julearn - INFO - Using dataframe as input
2021-01-28 20:06:58,159 - julearn - INFO - Features: ['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
2021-01-28 20:06:58,159 - julearn - INFO - Target: target
2021-01-28 20:06:58,159 - julearn - INFO - Expanded X: ['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
2021-01-28 20:06:58,160 - julearn - INFO - Expanded Confounds: []
2021-01-28 20:06:58,161 - julearn - INFO - ====================
2021-01-28 20:06:58,161 - julearn - INFO -
2021-01-28 20:06:58,161 - julearn - INFO - ====== Model ======
2021-01-28 20:06:58,161 - julearn - INFO - Obtaining model by name: ridge
2021-01-28 20:06:58,161 - julearn - INFO - ===================
2021-01-28 20:06:58,161 - julearn - INFO -
2021-01-28 20:06:58,161 - julearn - INFO - CV interpreted as RepeatedKFold with 5 repetitions of 5 folds
The scores dataframe has all the values for each CV split.
print(scores.head())
Out:
fit_time score_time test_neg_mean_absolute_error repeat fold
0 0.012433 0.007876 -53.711774 0 0
1 0.012493 0.007789 -38.334731 0 1
2 0.011724 0.007702 -46.578596 0 2
3 0.011671 0.007753 -48.109208 0 3
4 0.011724 0.007663 -42.632269 0 4
Mean value of mean absolute error across CV
print(scores['test_neg_mean_absolute_error'].mean() * -1)
Out:
45.94450832287054
Now we can get the MAE fold and repetition:
df_mae = scores.set_index(
['repeat', 'fold'])['test_neg_mean_absolute_error'].unstack() * -1
df_mae.index.name = 'Repeats'
df_mae.columns.name = 'K-fold splits'
print(df_mae)
Out:
K-fold splits 0 1 2 3 4
Repeats
0 53.711774 38.334731 46.578596 48.109208 42.632269
1 46.110360 49.273373 45.245755 48.425018 41.661824
2 45.176249 50.034589 48.108842 39.433303 44.937370
3 46.238680 39.120725 48.266876 46.853780 48.097210
4 45.943724 43.651725 41.527743 47.429889 53.709096
Plot heatmap of mean absolute error (MAE) over all repeats and CV splits
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.heatmap(df_mae, cmap="YlGnBu")
plt.title('Cross-validation MAE')
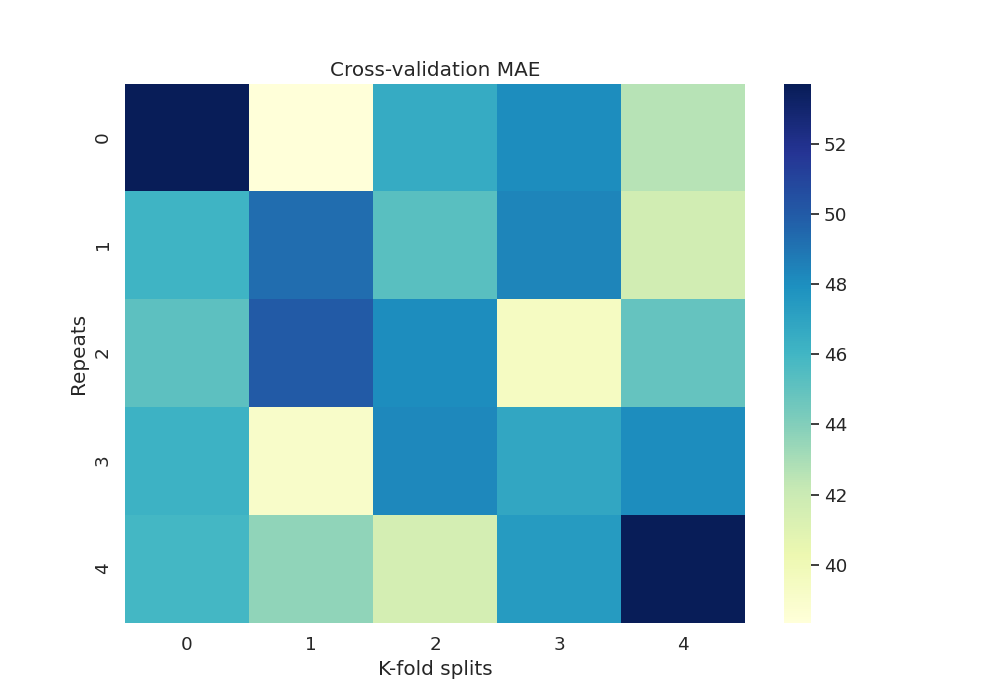
Out:
Text(0.5, 1.0, 'Cross-validation MAE')
Let’s plot the feature importance using the coefficients of the trained model
features = pd.DataFrame({'Features': X, 'importance': model['ridge'].coef_})
features.sort_values(by=['importance'], ascending=True, inplace=True)
features['positive'] = features['importance'] > 0
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
features.set_index('Features', inplace=True)
features.importance.plot(kind='barh',
color=features.positive.map
({True: 'blue', False: 'red'}))
ax.set(xlabel='Importance', title='Variable importance for Ridge Regression')
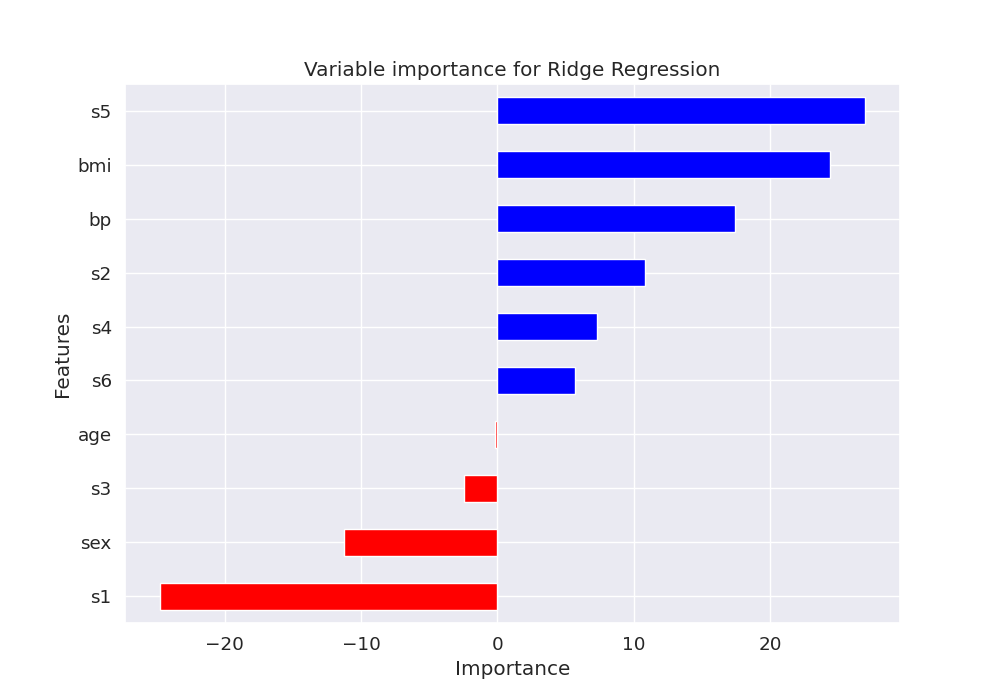
Out:
[Text(0.5, 40.249999999999986, 'Importance'), Text(0.5, 1.0, 'Variable importance for Ridge Regression')]
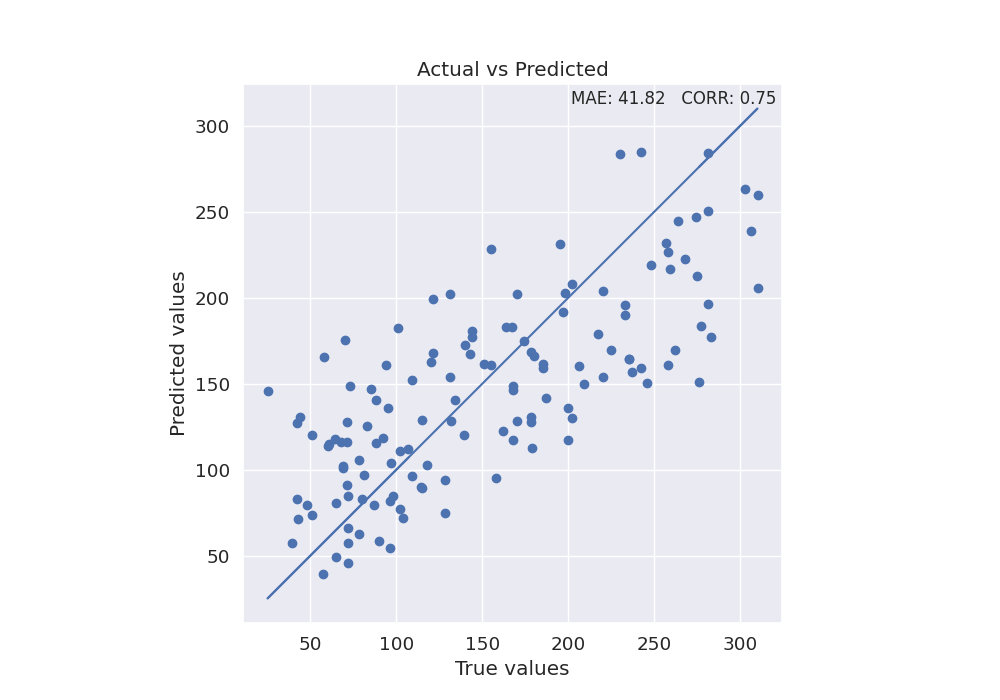
Use the final model to make predictions on test data and plot scatterplot of true values vs predicted values
y_true = test_diabetes[y]
y_pred = model.predict(test_diabetes[X])
mae = format(mean_absolute_error(y_true, y_pred), '.2f')
corr = format(np.corrcoef(y_pred, y_true)[1, 0], '.2f')
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
sns.set_style("darkgrid")
plt.scatter(y_true, y_pred)
plt.plot(y_true, y_true)
xmin, xmax = ax.get_xlim()
ymin, ymax = ax.get_ylim()
text = 'MAE: ' + str(mae) + ' CORR: ' + str(corr)
ax.set(xlabel='True values', ylabel='Predicted values')
plt.title('Actual vs Predicted')
plt.text(xmax - 0.01 * xmax, ymax - 0.01 * ymax, text, verticalalignment='top',
horizontalalignment='right', fontsize=12)
plt.axis('scaled')
Out:
(10.75, 324.25, 10.75, 324.25)
Total running time of the script: ( 0 minutes 1.492 seconds)